| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cAMP-specific 3',5'-cyclic phosphodiesterase 4A |
|---|
| Ligand | BDBM50393516 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_856424 (CHEMBL2161156) |
|---|
| IC50 | 2400±n/a nM |
|---|
| Citation |  Day, JP; Lindsay, B; Riddell, T; Jiang, Z; Allcock, RW; Abraham, A; Sookup, S; Christian, F; Bogum, J; Martin, EK; Rae, RL; Anthony, D; Rosair, GM; Houslay, DM; Huston, E; Baillie, GS; Klussmann, E; Houslay, MD; Adams, DR Elucidation of a structural basis for the inhibitor-driven, p62 (SQSTM1)-dependent intracellular redistribution of cAMP phosphodiesterase-4A4 (PDE4A4). J Med Chem54:3331-47 (2011) [PubMed] Article Day, JP; Lindsay, B; Riddell, T; Jiang, Z; Allcock, RW; Abraham, A; Sookup, S; Christian, F; Bogum, J; Martin, EK; Rae, RL; Anthony, D; Rosair, GM; Houslay, DM; Huston, E; Baillie, GS; Klussmann, E; Houslay, MD; Adams, DR Elucidation of a structural basis for the inhibitor-driven, p62 (SQSTM1)-dependent intracellular redistribution of cAMP phosphodiesterase-4A4 (PDE4A4). J Med Chem54:3331-47 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| cAMP-specific 3',5'-cyclic phosphodiesterase 4A |
|---|
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4A |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | DPDE2 | PDE46 | PDE4A | PDE4A_HUMAN | Phosphodiesterase 4 (PDE4) | Phosphodiesterase 4A | Phosphodiesterase 4A (PDE4) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 98113.27 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P27815 |
|---|
| Residue: | 886 |
|---|
| Sequence: | MEPPTVPSERSLSLSLPGPREGQATLKPPPQHLWRQPRTPIRIQQRGYSDSAERAERERQ
PHRPIERADAMDTSDRPGLRTTRMSWPSSFHGTGTGSGGAGGGSSRRFEAENGPTPSPGR
SPLDSQASPGLVLHAGAATSQRRESFLYRSDSDYDMSPKTMSRNSSVTSEAHAEDLIVTP
FAQVLASLRSVRSNFSLLTNVPVPSNKRSPLGGPTPVCKATLSEETCQQLARETLEELDW
CLEQLETMQTYRSVSEMASHKFKRMLNRELTHLSEMSRSGNQVSEYISTTFLDKQNEVEI
PSPTMKEREKQQAPRPRPSQPPPPPVPHLQPMSQITGLKKLMHSNSLNNSNIPRFGVKTD
QEELLAQELENLNKWGLNIFCVSDYAGGRSLTCIMYMIFQERDLLKKFRIPVDTMVTYML
TLEDHYHADVAYHNSLHAADVLQSTHVLLATPALDAVFTDLEILAALFAAAIHDVDHPGV
SNQFLINTNSELALMYNDESVLENHHLAVGFKLLQEDNCDIFQNLSKRQRQSLRKMVIDM
VLATDMSKHMTLLADLKTMVETKKVTSSGVLLLDNYSDRIQVLRNMVHCADLSNPTKPLE
LYRQWTDRIMAEFFQQGDRERERGMEISPMCDKHTASVEKSQVGFIDYIVHPLWETWADL
VHPDAQEILDTLEDNRDWYYSAIRQSPSPPPEEESRGPGHPPLPDKFQFELTLEEEEEEE
ISMAQIPCTAQEALTAQGLSGVEEALDATIAWEASPAQESLEVMAQEASLEAELEAVYLT
QQAQSTGSAPVAPDEFSSREEFVVAVSHSSPSALALQSPLLPAWRTLSVSEHAPGLPGLP
STAAEVEAQREHQAAKRACSACAGTFGEDTSALPAPGGGGSGGDPT
|
|
|
|---|
| BDBM50393516 |
|---|
| n/a |
|---|
| Name | BDBM50393516 |
|---|
| Synonyms: | CHEMBL2158062 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H23NO3 |
|---|
| Mol. Mass. | 289.3694 |
|---|
| SMILES | COc1ccc(cc1OC1CCCC1)[C@H]1CN(C)C(=O)C1 |r| |
|---|
| Structure | 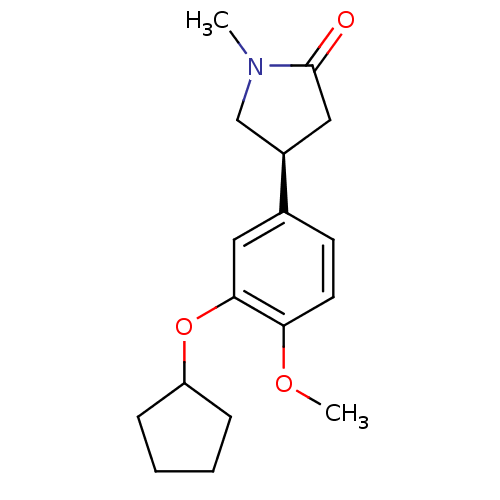
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Day, JP; Lindsay, B; Riddell, T; Jiang, Z; Allcock, RW; Abraham, A; Sookup, S; Christian, F; Bogum, J; Martin, EK; Rae, RL; Anthony, D; Rosair, GM; Houslay, DM; Huston, E; Baillie, GS; Klussmann, E; Houslay, MD; Adams, DR Elucidation of a structural basis for the inhibitor-driven, p62 (SQSTM1)-dependent intracellular redistribution of cAMP phosphodiesterase-4A4 (PDE4A4). J Med Chem54:3331-47 (2011) [PubMed] Article
Day, JP; Lindsay, B; Riddell, T; Jiang, Z; Allcock, RW; Abraham, A; Sookup, S; Christian, F; Bogum, J; Martin, EK; Rae, RL; Anthony, D; Rosair, GM; Houslay, DM; Huston, E; Baillie, GS; Klussmann, E; Houslay, MD; Adams, DR Elucidation of a structural basis for the inhibitor-driven, p62 (SQSTM1)-dependent intracellular redistribution of cAMP phosphodiesterase-4A4 (PDE4A4). J Med Chem54:3331-47 (2011) [PubMed] Article