| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Pyroglutamylated RF-amide peptide receptor |
|---|
| Ligand | BDBM50347818 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_862134 (CHEMBL2174319) |
|---|
| IC50 | 1.3±n/a nM |
|---|
| Citation |  Neveu, C; Lefranc, B; Tasseau, O; Do-Rego, JC; Bourmaud, A; Chan, P; Bauchat, P; Le Marec, O; Chuquet, J; Guilhaudis, L; Boutin, JA; Ségalas-Milazzo, I; Costentin, J; Vaudry, H; Baudy-Floc'h, M; Vaudry, D; Leprince, J Rational design of a low molecular weight, stable, potent, and long-lasting GPR103 aza-ß3-pseudopeptide agonist. J Med Chem55:7516-24 (2012) [PubMed] Article Neveu, C; Lefranc, B; Tasseau, O; Do-Rego, JC; Bourmaud, A; Chan, P; Bauchat, P; Le Marec, O; Chuquet, J; Guilhaudis, L; Boutin, JA; Ségalas-Milazzo, I; Costentin, J; Vaudry, H; Baudy-Floc'h, M; Vaudry, D; Leprince, J Rational design of a low molecular weight, stable, potent, and long-lasting GPR103 aza-ß3-pseudopeptide agonist. J Med Chem55:7516-24 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Pyroglutamylated RF-amide peptide receptor |
|---|
| Name: | Pyroglutamylated RF-amide peptide receptor |
|---|
| Synonyms: | GPR103 | Pyroglutamylated RFamide peptide receptor | QRFPR | QRFPR_HUMAN |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 49508.46 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | GPR103 0 HUMAN::Q96P65 |
|---|
| Residue: | 431 |
|---|
| Sequence: | MQALNITPEQFSRLLRDHNLTREQFIALYRLRPLVYTPELPGRAKLALVLTGVLIFALAL
FGNALVFYVVTRSKAMRTVTNIFICSLALSDLLITFFCIPVTMLQNISDNWLGGAFICKM
VPFVQSTAVVTEILTMTCIAVERHQGLVHPFKMKWQYTNRRAFTMLGVVWLVAVIVGSPM
WHVQQLEIKYDFLYEKEHICCLEEWTSPVHQKIYTTFILVILFLLPLMVMLILYSKIGYE
LWIKKRVGDGSVLRTIHGKEMSKIARKKKRAVIMMVTVVALFAVCWAPFHVVHMMIEYSN
FEKEYDDVTIKMIFAIVQIIGFSNSICNPIVYAFMNENFKKNVLSAVCYCIVNKTFSPAQ
RHGNSGITMMRKKAKFSLRENPVEETKGEAFSDGNIEVKLCEQTEEKKKLKRHLALFRSE
LAENSPLDSGH
|
|
|
|---|
| BDBM50347818 |
|---|
| n/a |
|---|
| Name | BDBM50347818 |
|---|
| Synonyms: | CHEMBL1802413 | P518 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C127H195N37O37 |
|---|
| Mol. Mass. | 2832.1329 |
|---|
| SMILES | [#6]-[#6](-[#6])-[#6]-[#6@H](-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6]-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#6])-[#6])-[#7]-[#6](=O)-[#6@@H]-1-[#6]-[#6]-[#6]-[#7]-1-[#6](=O)-[#6]-[#7]-[#6](=O)-[#6@H](-[#6]-[#8])-[#7]-[#6](=O)-[#6@@H](-[#7])-[#6@@H](-[#6])-[#8])-[#6](=O)-[#7]-[#6@@H](-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6](-[#6])-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6](-[#7])=O)-[#6](=O)-[#7]-[#6]-[#6](=O)-[#7]-[#6@@H](-[#6]-c1ccc(-[#8])cc1)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#8])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#6](=O)-[#7]-[#6]-[#6](=O)-[#7]-[#6]-[#6](=O)-[#7]-[#6@@H](-[#6]-c1ccccc1)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#8])-[#6](=O)-[#7]-[#6@@H](-[#6]-c1ccccc1)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-c1ccccc1)-[#6](-[#7])=O |r| |
|---|
| Structure | 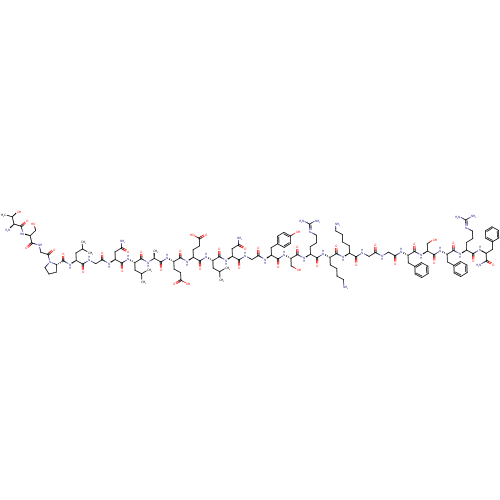
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Neveu, C; Lefranc, B; Tasseau, O; Do-Rego, JC; Bourmaud, A; Chan, P; Bauchat, P; Le Marec, O; Chuquet, J; Guilhaudis, L; Boutin, JA; Ségalas-Milazzo, I; Costentin, J; Vaudry, H; Baudy-Floc'h, M; Vaudry, D; Leprince, J Rational design of a low molecular weight, stable, potent, and long-lasting GPR103 aza-ß3-pseudopeptide agonist. J Med Chem55:7516-24 (2012) [PubMed] Article
Neveu, C; Lefranc, B; Tasseau, O; Do-Rego, JC; Bourmaud, A; Chan, P; Bauchat, P; Le Marec, O; Chuquet, J; Guilhaudis, L; Boutin, JA; Ségalas-Milazzo, I; Costentin, J; Vaudry, H; Baudy-Floc'h, M; Vaudry, D; Leprince, J Rational design of a low molecular weight, stable, potent, and long-lasting GPR103 aza-ß3-pseudopeptide agonist. J Med Chem55:7516-24 (2012) [PubMed] Article