| Reaction Details |
|---|
 | Report a problem with these data |
| Target | E3 ubiquitin-protein ligase Mdm2 |
|---|
| Ligand | BDBM50388626 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_874354 (CHEMBL2182652) |
|---|
| Kd | 0.4±n/a nM |
|---|
| Citation |  Rew, Y; Sun, D; Gonzalez-Lopez De Turiso, F; Bartberger, MD; Beck, HP; Canon, J; Chen, A; Chow, D; Deignan, J; Fox, BM; Gustin, D; Huang, X; Jiang, M; Jiao, X; Jin, L; Kayser, F; Kopecky, DJ; Li, Y; Lo, MC; Long, AM; Michelsen, K; Oliner, JD; Osgood, T; Ragains, M; Saiki, AY; Schneider, S; Toteva, M; Yakowec, P; Yan, X; Ye, Q; Yu, D; Zhao, X; Zhou, J; Medina, JC; Olson, SH Structure-based design of novel inhibitors of the MDM2-p53 interaction. J Med Chem55:4936-54 (2012) [PubMed] Article Rew, Y; Sun, D; Gonzalez-Lopez De Turiso, F; Bartberger, MD; Beck, HP; Canon, J; Chen, A; Chow, D; Deignan, J; Fox, BM; Gustin, D; Huang, X; Jiang, M; Jiao, X; Jin, L; Kayser, F; Kopecky, DJ; Li, Y; Lo, MC; Long, AM; Michelsen, K; Oliner, JD; Osgood, T; Ragains, M; Saiki, AY; Schneider, S; Toteva, M; Yakowec, P; Yan, X; Ye, Q; Yu, D; Zhao, X; Zhou, J; Medina, JC; Olson, SH Structure-based design of novel inhibitors of the MDM2-p53 interaction. J Med Chem55:4936-54 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| E3 ubiquitin-protein ligase Mdm2 |
|---|
| Name: | E3 ubiquitin-protein ligase Mdm2 |
|---|
| Synonyms: | Double minute 2 protein | Double minute 2 protein (HDM2) | E3 ubiquitin-protein ligase Mdm2 (p53-binding protein Mdm2) | Hdm2 | Human Double Minute 2 (HDM2) | MDM2 | MDM2-MDMX | MDM2_HUMAN | p53-Binding Protein MDM2 | p53-binding protein |
|---|
| Type: | Oncoprotein |
|---|
| Mol. Mass.: | 55196.54 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q00987 |
|---|
| Residue: | 491 |
|---|
| Sequence: | MCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQY
IMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGT
SVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRRRAISETEENSDELSGERQ
RKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDS
VSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEVYQVTVYQAGESDTDSFEEDPEISLA
DYWKCTSCNEMNPPLPSHCNRCWALRENWLPEDKGKDKGEISEKAKLENSTQAEEGFDVP
DCKKTIVNDSRESCVEENDDKITQASQSQESEDYSQPSTSSSIIYSSQEDVKEFEREETQ
DKEESVESSLPLNAIEPCVICQGRPKNGCIVHGKTGHLMACFTCAKKLKKRNKPCPVCRQ
PIQMIVLTYFP
|
|
|
|---|
| BDBM50388626 |
|---|
| n/a |
|---|
| Name | BDBM50388626 |
|---|
| Synonyms: | CHEMBL2059435 | US9296736, 152 | US9593129, Example 152 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H29Cl2NO4 |
|---|
| Mol. Mass. | 478.408 |
|---|
| SMILES | CC[C@@H]([C@H](C)O)N1[C@@H]([C@H](C[C@](C)(CC(O)=O)C1=O)c1cccc(Cl)c1)c1ccc(Cl)cc1 |r| |
|---|
| Structure | 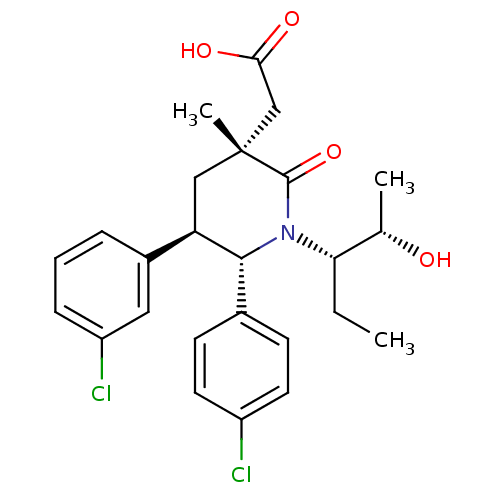
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Rew, Y; Sun, D; Gonzalez-Lopez De Turiso, F; Bartberger, MD; Beck, HP; Canon, J; Chen, A; Chow, D; Deignan, J; Fox, BM; Gustin, D; Huang, X; Jiang, M; Jiao, X; Jin, L; Kayser, F; Kopecky, DJ; Li, Y; Lo, MC; Long, AM; Michelsen, K; Oliner, JD; Osgood, T; Ragains, M; Saiki, AY; Schneider, S; Toteva, M; Yakowec, P; Yan, X; Ye, Q; Yu, D; Zhao, X; Zhou, J; Medina, JC; Olson, SH Structure-based design of novel inhibitors of the MDM2-p53 interaction. J Med Chem55:4936-54 (2012) [PubMed] Article
Rew, Y; Sun, D; Gonzalez-Lopez De Turiso, F; Bartberger, MD; Beck, HP; Canon, J; Chen, A; Chow, D; Deignan, J; Fox, BM; Gustin, D; Huang, X; Jiang, M; Jiao, X; Jin, L; Kayser, F; Kopecky, DJ; Li, Y; Lo, MC; Long, AM; Michelsen, K; Oliner, JD; Osgood, T; Ragains, M; Saiki, AY; Schneider, S; Toteva, M; Yakowec, P; Yan, X; Ye, Q; Yu, D; Zhao, X; Zhou, J; Medina, JC; Olson, SH Structure-based design of novel inhibitors of the MDM2-p53 interaction. J Med Chem55:4936-54 (2012) [PubMed] Article