

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Corticotropin-releasing factor receptor 1 | ||
| Ligand | BDBM50398587 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_873519 (CHEMBL2187021) | ||
| IC50 | 46±n/a nM | ||
| Citation |  Takahashi, Y; Hashizume, M; Shin, K; Terauchi, T; Takeda, K; Hibi, S; Murata-Tai, K; Fujisawa, M; Shikata, K; Taguchi, R; Ino, M; Shibata, H; Yonaga, M Design, synthesis, and structure-activity relationships of novel pyrazolo[5,1-b]thiazole derivatives as potent and orally active corticotropin-releasing factor 1 receptor antagonists. J Med Chem55:8450-63 (2012) [PubMed] Article Takahashi, Y; Hashizume, M; Shin, K; Terauchi, T; Takeda, K; Hibi, S; Murata-Tai, K; Fujisawa, M; Shikata, K; Taguchi, R; Ino, M; Shibata, H; Yonaga, M Design, synthesis, and structure-activity relationships of novel pyrazolo[5,1-b]thiazole derivatives as potent and orally active corticotropin-releasing factor 1 receptor antagonists. J Med Chem55:8450-63 (2012) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Corticotropin-releasing factor receptor 1 | |||
| Name: | Corticotropin-releasing factor receptor 1 | ||
| Synonyms: | CRF-R | CRF-R2 Alpha | CRF1 | CRFR | CRFR1 | CRFR1_HUMAN | CRH-R 1 | CRHR | CRHR1 | Corticotropin releasing factor receptor 1 | Corticotropin-releasing factor receptor 1 (CRF-1) | Corticotropin-releasing factor receptor 1 (CRF1) | Corticotropin-releasing hormone receptor 1 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 50744.31 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P34998 | ||
| Residue: | 444 | ||
| Sequence: |
| ||
| BDBM50398587 | |||
| n/a | |||
| Name | BDBM50398587 | ||
| Synonyms: | CHEMBL2179197 | US8530504, 4 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H35N3O4S | ||
| Mol. Mass. | 485.639 | ||
| SMILES | COCc1cc(OC)c(-c2csc3c(N(CC4CC4)CC4CCOCC4)c(C)nn23)c(OC)c1 |(17.24,-37.02,;16.5,-35.68,;14.96,-35.65,;14.21,-34.3,;12.67,-34.28,;11.93,-32.93,;10.39,-32.91,;9.59,-34.23,;12.71,-31.61,;11.97,-30.27,;12.84,-29.01,;11.92,-27.78,;10.46,-28.29,;8.99,-27.85,;8.44,-26.4,;6.92,-26.15,;5.95,-27.34,;4.51,-27.89,;5.71,-28.86,;9.2,-25.06,;8.41,-23.74,;9.16,-22.4,;8.37,-21.07,;6.83,-21.09,;6.08,-22.44,;6.87,-23.76,;8.11,-29.11,;6.57,-29.14,;9.04,-30.33,;10.5,-29.82,;14.25,-31.64,;15.05,-30.32,;16.58,-30.34,;15,-32.99,)| | ||
| Structure | 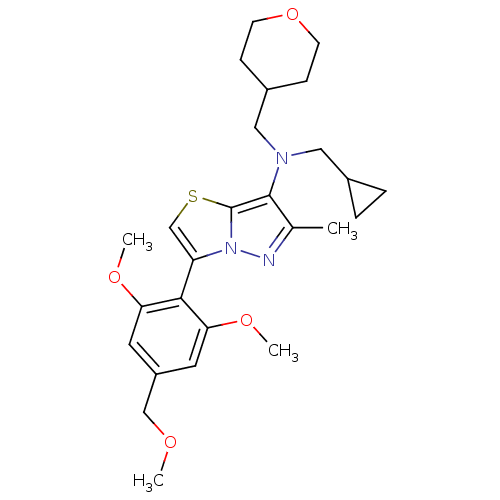
| ||