| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Polyamine deacetylase HDAC10 |
|---|
| Ligand | BDBM50398716 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_875949 (CHEMBL2188455) |
|---|
| IC50 | 7570±n/a nM |
|---|
| Citation |  Bergman, JA; Woan, K; Perez-Villarroel, P; Villagra, A; Sotomayor, EM; Kozikowski, AP Selective histone deacetylase 6 inhibitors bearing substituted urea linkers inhibit melanoma cell growth. J Med Chem55:9891-9 (2012) [PubMed] Article Bergman, JA; Woan, K; Perez-Villarroel, P; Villagra, A; Sotomayor, EM; Kozikowski, AP Selective histone deacetylase 6 inhibitors bearing substituted urea linkers inhibit melanoma cell growth. J Med Chem55:9891-9 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Polyamine deacetylase HDAC10 |
|---|
| Name: | Polyamine deacetylase HDAC10 |
|---|
| Synonyms: | HD10 | HDA10_HUMAN | HDAC10 | Histone deacetylase | Histone deacetylase 10 | Human HDAC10 |
|---|
| Type: | Chromatin regulator; hydrolase; repressor |
|---|
| Mol. Mass.: | 71431.89 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q969S8 |
|---|
| Residue: | 669 |
|---|
| Sequence: | MGTALVYHEDMTATRLLWDDPECEIERPERLTAALDRLRQRGLEQRCLRLSAREASEEEL
GLVHSPEYVSLVRETQVLGKEELQALSGQFDAIYFHPSTFHCARLAAGAGLQLVDAVLTG
AVQNGLALVRPPGHHGQRAAANGFCVFNNVAIAAAHAKQKHGLHRILVVDWDVHHGQGIQ
YLFEDDPSVLYFSWHRYEHGRFWPFLRESDADAVGRGQGLGFTVNLPWNQVGMGNADYVA
AFLHLLLPLAFEFDPELVLVSAGFDSAIGDPEGQMQATPECFAHLTQLLQVLAGGRVCAV
LEGGYHLESLAESVCMTVQTLLGDPAPPLSGPMAPCQSALESIQSARAAQAPHWKSLQQQ
DVTAVPMSPSSHSPEGRPPPLLPGGPVCKAAASAPSSLLDQPCLCPAPSVRTAVALTTPD
ITLVLPPDVIQQEASALREETEAWARPHESLAREEALTALGKLLYLLDGMLDGQVNSGIA
ATPASAAAATLDVAVRRGLSHGAQRLLCVALGQLDRPPDLAHDGRSLWLNIRGKEAAALS
MFHVSTPLPVMTGGFLSCILGLVLPLAYGFQPDLVLVALGPGHGLQGPHAALLAAMLRGL
AGGRVLALLEENSTPQLAGILARVLNGEAPPSLGPSSVASPEDVQALMYLRGQLEPQWKM
LQCHPHLVA
|
|
|
|---|
| BDBM50398716 |
|---|
| n/a |
|---|
| Name | BDBM50398716 |
|---|
| Synonyms: | CHEMBL2179618 | US10227295, Compound 5g | US9409858, 5g | US9751832, Compound 5g | US9956192, Compound 5g |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H23N3O3 |
|---|
| Mol. Mass. | 341.4042 |
|---|
| SMILES | CCCCN(Cc1ccc(cc1)C(=O)NO)C(=O)Nc1ccccc1 |
|---|
| Structure | 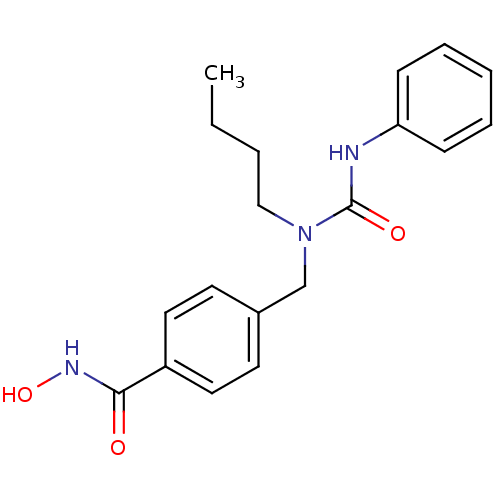
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Bergman, JA; Woan, K; Perez-Villarroel, P; Villagra, A; Sotomayor, EM; Kozikowski, AP Selective histone deacetylase 6 inhibitors bearing substituted urea linkers inhibit melanoma cell growth. J Med Chem55:9891-9 (2012) [PubMed] Article
Bergman, JA; Woan, K; Perez-Villarroel, P; Villagra, A; Sotomayor, EM; Kozikowski, AP Selective histone deacetylase 6 inhibitors bearing substituted urea linkers inhibit melanoma cell growth. J Med Chem55:9891-9 (2012) [PubMed] Article