| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 3-phosphoinositide-dependent protein kinase 1 |
|---|
| Ligand | BDBM50399645 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_878678 (CHEMBL2182502) |
|---|
| Kd | 43±n/a nM |
|---|
| Citation |  Wilhelm, A; Lopez-Garcia, LA; Busschots, K; Fröhner, W; Maurer, F; Boettcher, S; Zhang, H; Schulze, JO; Biondi, RM; Engel, M 2-(3-Oxo-1,3-diphenylpropyl)malonic acids as potent allosteric ligands of the PIF pocket of phosphoinositide-dependent kinase-1: development and prodrug concept. J Med Chem55:9817-30 (2012) [PubMed] Article Wilhelm, A; Lopez-Garcia, LA; Busschots, K; Fröhner, W; Maurer, F; Boettcher, S; Zhang, H; Schulze, JO; Biondi, RM; Engel, M 2-(3-Oxo-1,3-diphenylpropyl)malonic acids as potent allosteric ligands of the PIF pocket of phosphoinositide-dependent kinase-1: development and prodrug concept. J Med Chem55:9817-30 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 3-phosphoinositide-dependent protein kinase 1 |
|---|
| Name: | 3-phosphoinositide-dependent protein kinase 1 |
|---|
| Synonyms: | 3-Phosphoinositide-Dependent Protein Kinase 1 (PDK1) | 3-phosphoinositide dependent protein kinase-1 | 3-phosphoinositide-dependent protein kinase 1 | 3-phosphoinositide-dependent protein kinase 1 (PDK) | 3-phosphoinositide-dependent protein kinase 1 (PDK-1) | 3-phosphoinositide-dependent protein kinase 1 (PDK1)(Δ1-50) | Isoform 2 of 3-phosphoinositide-dependent protein kinase 1 | PDK1 | PDPK1 | PDPK1_HUMAN | Phosphoinositide-dependent protein kinase 1 (PDK1) | Pyruvate dehydrogenase kinase isoenzyme 1 (PDK1) | hPDK1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 63157.65 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15530 |
|---|
| Residue: | 556 |
|---|
| Sequence: | MARTTSQLYDAVPIQSSVVLCSCPSPSMVRTQTESSTPPGIPGGSRQGPAMDGTAAEPRP
GAGSLQHAQPPPQPRKKRPEDFKFGKILGEGSFSTVVLARELATSREYAIKILEKRHIIK
ENKVPYVTRERDVMSRLDHPFFVKLYFTFQDDEKLYFGLSYAKNGELLKYIRKIGSFDET
CTRFYTAEIVSALEYLHGKGIIHRDLKPENILLNEDMHIQITDFGTAKVLSPESKQARAN
SFVGTAQYVSPELLTEKSACKSSDLWALGCIIYQLVAGLPPFRAGNEYLIFQKIIKLEYD
FPEKFFPKARDLVEKLLVLDATKRLGCEEMEGYGPLKAHPFFESVTWENLHQQTPPKLTA
YLPAMSEDDEDCYGNYDNLLSQFGCMQVSSSSSSHSLSASDTGLPQRSGSNIEQYIHDLD
SNSFELDLQFSEDEKRLLLEKQAGGNPWHQFVENNLILKMGPVDKRKGLFARRRQLLLTE
GPHLYYVDPVNKVLKGEIPWSQELRPEAKNFKTFFVHTPNRTYYLMDPSGNAHKWCRKIQ
EVWRQRYQSHPDAAVQ
|
|
|
|---|
| BDBM50399645 |
|---|
| n/a |
|---|
| Name | BDBM50399645 |
|---|
| Synonyms: | CHEMBL1213203 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C135H197N35O44S2 |
|---|
| Mol. Mass. | 3078.347 |
|---|
| SMILES | CC[C@H](C)[C@H](NC(=O)[C@H](Cc1ccc(O)cc1)NC(=O)[C@H](CC(O)=O)NC(=O)[C@H](Cc1ccccc1)NC(=O)[C@H](CC(O)=O)NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@H](Cc1ccccc1)NC(=O)[C@H](CCSC)NC(=O)[C@H](CCC(O)=O)NC(=O)[C@H](CCC(N)=O)NC(=O)[C@H](CCC(O)=O)NC(=O)[C@H](CCC(O)=O)NC(=O)[C@H](CCC(O)=O)NC(=O)[C@H](CO)NC(=O)[C@H](CC(C)C)NC(=O)[C@@H](NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@@H]1CCCN1C(=O)[C@H](CCC(O)=O)NC(=O)[C@@H](N)CCCNC(N)=N)[C@@H](C)CC)C(=O)N[C@@H](C)C(=O)N[C@@H](CC(O)=O)C(=O)N[C@@H](Cc1c[nH]c2ccccc12)C(=O)N[C@@H](CS)C(O)=O |r| |
|---|
| Structure | 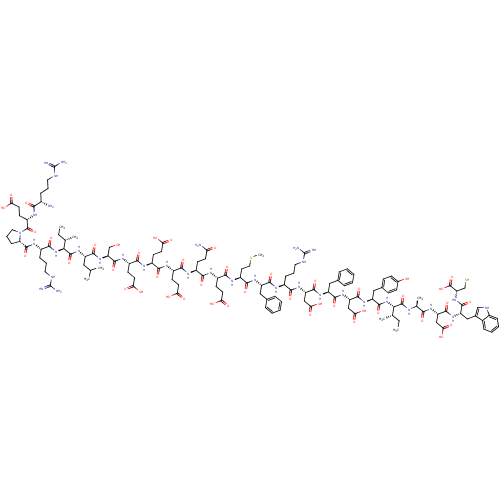
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wilhelm, A; Lopez-Garcia, LA; Busschots, K; Fröhner, W; Maurer, F; Boettcher, S; Zhang, H; Schulze, JO; Biondi, RM; Engel, M 2-(3-Oxo-1,3-diphenylpropyl)malonic acids as potent allosteric ligands of the PIF pocket of phosphoinositide-dependent kinase-1: development and prodrug concept. J Med Chem55:9817-30 (2012) [PubMed] Article
Wilhelm, A; Lopez-Garcia, LA; Busschots, K; Fröhner, W; Maurer, F; Boettcher, S; Zhang, H; Schulze, JO; Biondi, RM; Engel, M 2-(3-Oxo-1,3-diphenylpropyl)malonic acids as potent allosteric ligands of the PIF pocket of phosphoinositide-dependent kinase-1: development and prodrug concept. J Med Chem55:9817-30 (2012) [PubMed] Article