| Reaction Details |
|---|
 | Report a problem with these data |
| Target | MAP kinase-interacting serine/threonine-protein kinase 2 |
|---|
| Ligand | BDBM50399676 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_876561 (CHEMBL2188518) |
|---|
| IC50 | 3200±n/a nM |
|---|
| Citation |  Yang, J; Wang, LJ; Liu, JJ; Zhong, L; Zheng, RL; Xu, Y; Ji, P; Zhang, CH; Wang, WJ; Lin, XD; Li, LL; Wei, YQ; Yang, SY Structural optimization and structure-activity relationships of N2-(4-(4-Methylpiperazin-1-yl)phenyl)-N8-phenyl-9H-purine-2,8-diamine derivatives, a new class of reversible kinase inhibitors targeting both EGFR-activating and resistance mutations. J Med Chem55:10685-99 (2012) [PubMed] Article Yang, J; Wang, LJ; Liu, JJ; Zhong, L; Zheng, RL; Xu, Y; Ji, P; Zhang, CH; Wang, WJ; Lin, XD; Li, LL; Wei, YQ; Yang, SY Structural optimization and structure-activity relationships of N2-(4-(4-Methylpiperazin-1-yl)phenyl)-N8-phenyl-9H-purine-2,8-diamine derivatives, a new class of reversible kinase inhibitors targeting both EGFR-activating and resistance mutations. J Med Chem55:10685-99 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| MAP kinase-interacting serine/threonine-protein kinase 2 |
|---|
| Name: | MAP kinase-interacting serine/threonine-protein kinase 2 |
|---|
| Synonyms: | GPRK7 | MAP kinase signal-integrating kinase 2 | MAP kinase-interacting serine/threonine-protein kinase 2 (MKNK2) | MAP kinase-interacting serine/threonine-protein kinase 2 (MnK2) | MAP-kinase interacting kinase 2 (MNK2) | MKNK2 | MKNK2_HUMAN | MNK2 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 51870.79 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9HBH9 |
|---|
| Residue: | 465 |
|---|
| Sequence: | MVQKKPAELQGFHRSFKGQNPFELAFSLDQPDHGDSDFGLQCSARPDMPASQPIDIPDAK
KRGKKKKRGRATDSFSGRFEDVYQLQEDVLGEGAHARVQTCINLITSQEYAVKIIEKQPG
HIRSRVFREVEMLYQCQGHRNVLELIEFFEEEDRFYLVFEKMRGGSILSHIHKRRHFNEL
EASVVVQDVASALDFLHNKGIAHRDLKPENILCEHPNQVSPVKICDFDLGSGIKLNGDCS
PISTPELLTPCGSAEYMAPEVVEAFSEEASIYDKRCDLWSLGVILYILLSGYPPFVGRCG
SDCGWDRGEACPACQNMLFESIQEGKYEFPDKDWAHISCAAKDLISKLLVRDAKQRLSAA
QVLQHPWVQGCAPENTLPTPMVLQRNSCAKDLTSFAAEAIAMNRQLAQHDEDLAEEEAAG
QGQPVLVRATSRCLQLSPPSQSKLAQRRQRASLSSAPVVLVGDHA
|
|
|
|---|
| BDBM50399676 |
|---|
| n/a |
|---|
| Name | BDBM50399676 |
|---|
| Synonyms: | CHEMBL2178352 | US9096601, 8-26 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H32N8 |
|---|
| Mol. Mass. | 468.5966 |
|---|
| SMILES | CN1CCN(CC1)c1ccc(Nc2ncc3nc(Nc4ccccc4)n(C4CCCC4)c3n2)cc1 |
|---|
| Structure | 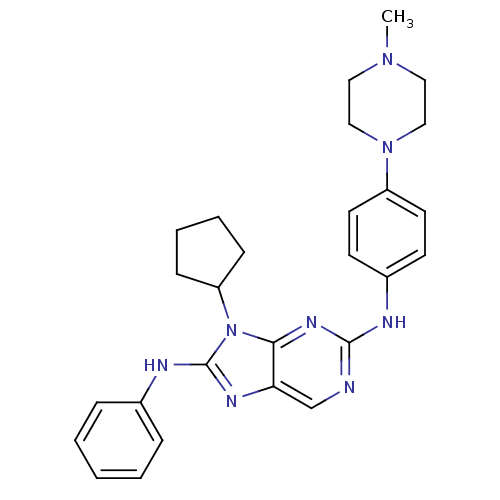
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Yang, J; Wang, LJ; Liu, JJ; Zhong, L; Zheng, RL; Xu, Y; Ji, P; Zhang, CH; Wang, WJ; Lin, XD; Li, LL; Wei, YQ; Yang, SY Structural optimization and structure-activity relationships of N2-(4-(4-Methylpiperazin-1-yl)phenyl)-N8-phenyl-9H-purine-2,8-diamine derivatives, a new class of reversible kinase inhibitors targeting both EGFR-activating and resistance mutations. J Med Chem55:10685-99 (2012) [PubMed] Article
Yang, J; Wang, LJ; Liu, JJ; Zhong, L; Zheng, RL; Xu, Y; Ji, P; Zhang, CH; Wang, WJ; Lin, XD; Li, LL; Wei, YQ; Yang, SY Structural optimization and structure-activity relationships of N2-(4-(4-Methylpiperazin-1-yl)phenyl)-N8-phenyl-9H-purine-2,8-diamine derivatives, a new class of reversible kinase inhibitors targeting both EGFR-activating and resistance mutations. J Med Chem55:10685-99 (2012) [PubMed] Article