| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Macrophage-stimulating protein receptor |
|---|
| Ligand | BDBM50399676 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_876729 (CHEMBL2183200) |
|---|
| IC50 | >10000±n/a nM |
|---|
| Citation |  Yang, J; Wang, LJ; Liu, JJ; Zhong, L; Zheng, RL; Xu, Y; Ji, P; Zhang, CH; Wang, WJ; Lin, XD; Li, LL; Wei, YQ; Yang, SY Structural optimization and structure-activity relationships of N2-(4-(4-Methylpiperazin-1-yl)phenyl)-N8-phenyl-9H-purine-2,8-diamine derivatives, a new class of reversible kinase inhibitors targeting both EGFR-activating and resistance mutations. J Med Chem55:10685-99 (2012) [PubMed] Article Yang, J; Wang, LJ; Liu, JJ; Zhong, L; Zheng, RL; Xu, Y; Ji, P; Zhang, CH; Wang, WJ; Lin, XD; Li, LL; Wei, YQ; Yang, SY Structural optimization and structure-activity relationships of N2-(4-(4-Methylpiperazin-1-yl)phenyl)-N8-phenyl-9H-purine-2,8-diamine derivatives, a new class of reversible kinase inhibitors targeting both EGFR-activating and resistance mutations. J Med Chem55:10685-99 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Macrophage-stimulating protein receptor |
|---|
| Name: | Macrophage-stimulating protein receptor |
|---|
| Synonyms: | 2.7.10.1 | CD_antigen=CD136 | CDw136 | MSP receptor | MST1R | Macrophage-stimulating protein receptor (MST1R) | Macrophage-stimulating protein receptor alpha chain | Macrophage-stimulating protein receptor beta chain | PTK8 | Protein-tyrosine kinase 8 | RON | RON_HUMAN | Tyrosine kinase receptor ron | p185-Ron |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 152270.76 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q04912 |
|---|
| Residue: | 1400 |
|---|
| Sequence: | MELLPPLPQSFLLLLLLPAKPAAGEDWQCPRTPYAASRDFDVKYVVPSFSAGGLVQAMVT
YEGDRNESAVFVAIRNRLHVLGPDLKSVQSLATGPAGDPGCQTCAACGPGPHGPPGDTDT
KVLVLDPALPALVSCGSSLQGRCFLHDLEPQGTAVHLAAPACLFSAHHNRPDDCPDCVAS
PLGTRVTVVEQGQASYFYVASSLDAAVAASFSPRSVSIRRLKADASGFAPGFVALSVLPK
HLVSYSIEYVHSFHTGAFVYFLTVQPASVTDDPSALHTRLARLSATEPELGDYRELVLDC
RFAPKRRRRGAPEGGQPYPVLRVAHSAPVGAQLATELSIAEGQEVLFGVFVTGKDGGPGV
GPNSVVCAFPIDLLDTLIDEGVERCCESPVHPGLRRGLDFFQSPSFCPNPPGLEALSPNT
SCRHFPLLVSSSFSRVDLFNGLLGPVQVTALYVTRLDNVTVAHMGTMDGRILQVELVRSL
NYLLYVSNFSLGDSGQPVQRDVSRLGDHLLFASGDQVFQVPIQGPGCRHFLTCGRCLRAW
HFMGCGWCGNMCGQQKECPGSWQQDHCPPKLTEFHPHSGPLRGSTRLTLCGSNFYLHPSG
LVPEGTHQVTVGQSPCRPLPKDSSKLRPVPRKDFVEEFECELEPLGTQAVGPTNVSLTVT
NMPPGKHFRVDGTSVLRGFSFMEPVLIAVQPLFGPRAGGTCLTLEGQSLSVGTSRAVLVN
GTECLLARVSEGQLLCATPPGATVASVPLSLQVGGAQVPGSWTFQYREDPVVLSISPNCG
YINSHITICGQHLTSAWHLVLSFHDGLRAVESRCERQLPEQQLCRLPEYVVRDPQGWVAG
NLSARGDGAAGFTLPGFRFLPPPHPPSANLVPLKPEEHAIKFEYIGLGAVADCVGINVTV
GGESCQHEFRGDMVVCPLPPSLQLGQDGAPLQVCVDGECHILGRVVRPGPDGVPQSTLLG
ILLPLLLLVAALATALVFSYWWRRKQLVLPPNLNDLASLDQTAGATPLPILYSGSDYRSG
LALPAIDGLDSTTCVHGASFSDSEDESCVPLLRKESIQLRDLDSALLAEVKDVLIPHERV
VTHSDRVIGKGHFGVVYHGEYIDQAQNRIQCAIKSLSRITEMQQVEAFLREGLLMRGLNH
PNVLALIGIMLPPEGLPHVLLPYMCHGDLLQFIRSPQRNPTVKDLISFGLQVARGMEYLA
EQKFVHRDLAARNCMLDESFTVKVADFGLARDILDREYYSVQQHRHARLPVKWMALESLQ
TYRFTTKSDVWSFGVLLWELLTRGAPPYRHIDPFDLTHFLAQGRRLPQPEYCPDSLYQVM
QQCWEADPAVRPTFRVLVGEVEQIVSALLGDHYVQLPATYMNLGPSTSHEMNVRPEQPQF
SPMPGNVRRPRPLSEPPRPT
|
|
|
|---|
| BDBM50399676 |
|---|
| n/a |
|---|
| Name | BDBM50399676 |
|---|
| Synonyms: | CHEMBL2178352 | US9096601, 8-26 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H32N8 |
|---|
| Mol. Mass. | 468.5966 |
|---|
| SMILES | CN1CCN(CC1)c1ccc(Nc2ncc3nc(Nc4ccccc4)n(C4CCCC4)c3n2)cc1 |
|---|
| Structure | 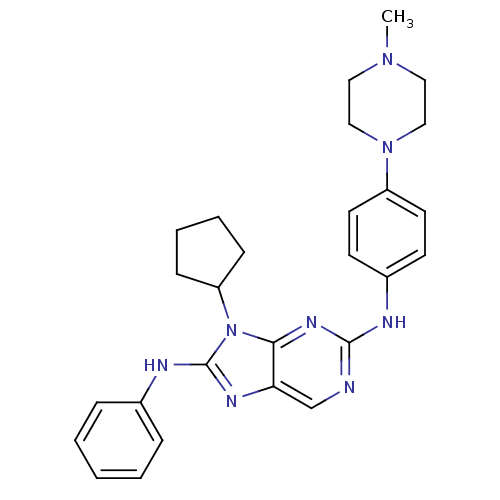
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Yang, J; Wang, LJ; Liu, JJ; Zhong, L; Zheng, RL; Xu, Y; Ji, P; Zhang, CH; Wang, WJ; Lin, XD; Li, LL; Wei, YQ; Yang, SY Structural optimization and structure-activity relationships of N2-(4-(4-Methylpiperazin-1-yl)phenyl)-N8-phenyl-9H-purine-2,8-diamine derivatives, a new class of reversible kinase inhibitors targeting both EGFR-activating and resistance mutations. J Med Chem55:10685-99 (2012) [PubMed] Article
Yang, J; Wang, LJ; Liu, JJ; Zhong, L; Zheng, RL; Xu, Y; Ji, P; Zhang, CH; Wang, WJ; Lin, XD; Li, LL; Wei, YQ; Yang, SY Structural optimization and structure-activity relationships of N2-(4-(4-Methylpiperazin-1-yl)phenyl)-N8-phenyl-9H-purine-2,8-diamine derivatives, a new class of reversible kinase inhibitors targeting both EGFR-activating and resistance mutations. J Med Chem55:10685-99 (2012) [PubMed] Article