

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Diacylglycerol O-acyltransferase 1 | ||
| Ligand | BDBM50399700 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_876789 (CHEMBL2183606) | ||
| IC50 | >10000±n/a nM | ||
| Citation |  Barlind, JG; Bauer, UA; Birch, AM; Birtles, S; Buckett, LK; Butlin, RJ; Davies, RD; Eriksson, JW; Hammond, CD; Hovland, R; Johannesson, P; Johansson, MJ; Kemmitt, PD; Lindmark, BT; Morentin Gutierrez, P; Noeske, TA; Nordin, A; O'Donnell, CJ; Petersson, AU; Redzic, A; Turnbull, AV; Vinblad, J Design and optimization of pyrazinecarboxamide-based inhibitors of diacylglycerol acyltransferase 1 (DGAT1) leading to a clinical candidate dimethylpyrazinecarboxamide phenylcyclohexylacetic acid (AZD7687). J Med Chem55:10610-29 (2012) [PubMed] Article Barlind, JG; Bauer, UA; Birch, AM; Birtles, S; Buckett, LK; Butlin, RJ; Davies, RD; Eriksson, JW; Hammond, CD; Hovland, R; Johannesson, P; Johansson, MJ; Kemmitt, PD; Lindmark, BT; Morentin Gutierrez, P; Noeske, TA; Nordin, A; O'Donnell, CJ; Petersson, AU; Redzic, A; Turnbull, AV; Vinblad, J Design and optimization of pyrazinecarboxamide-based inhibitors of diacylglycerol acyltransferase 1 (DGAT1) leading to a clinical candidate dimethylpyrazinecarboxamide phenylcyclohexylacetic acid (AZD7687). J Med Chem55:10610-29 (2012) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Diacylglycerol O-acyltransferase 1 | |||
| Name: | Diacylglycerol O-acyltransferase 1 | ||
| Synonyms: | ACAT-related gene product 1 | AGRP1 | Acyl coA-diacylglycerol acyl transferase 1 (DGAT1) | DGAT | DGAT1 | DGAT1_HUMAN | Diacylglycerol Acyltransferase (DGAT1) | Diacylglycerol O-acyltransferase 1 | Diacylglycerol O-acyltransferase 1 (DGAT1) | Diglyceride acyltransferase | ||
| Type: | Protein | ||
| Mol. Mass.: | 55297.82 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | O75907 | ||
| Residue: | 488 | ||
| Sequence: |
| ||
| BDBM50399700 | |||
| n/a | |||
| Name | BDBM50399700 | ||
| Synonyms: | CHEMBL2178391 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H27N3O3 | ||
| Mol. Mass. | 429.5109 | ||
| SMILES | Cc1nc(-c2ccccc2)c(nc1-c1ccc(cc1)[C@H]1CC[C@H](CC(O)=O)CC1)C(N)=O |r,wU:19.21,wD:22.25,(28.61,-10.63,;27.27,-9.87,;25.94,-10.64,;24.6,-9.87,;23.28,-10.64,;21.94,-9.87,;20.61,-10.64,;20.61,-12.18,;21.95,-12.95,;23.28,-12.18,;24.6,-8.33,;25.94,-7.56,;27.27,-8.33,;28.6,-7.56,;29.94,-8.34,;31.27,-7.56,;31.27,-6.01,;29.93,-5.25,;28.6,-6.02,;32.6,-5.24,;32.59,-3.7,;33.93,-2.93,;35.27,-3.7,;36.6,-2.93,;37.93,-3.7,;39.27,-2.93,;37.93,-5.24,;35.26,-5.24,;33.94,-6.01,;23.27,-7.56,;21.94,-8.32,;23.27,-6.02,)| | ||
| Structure | 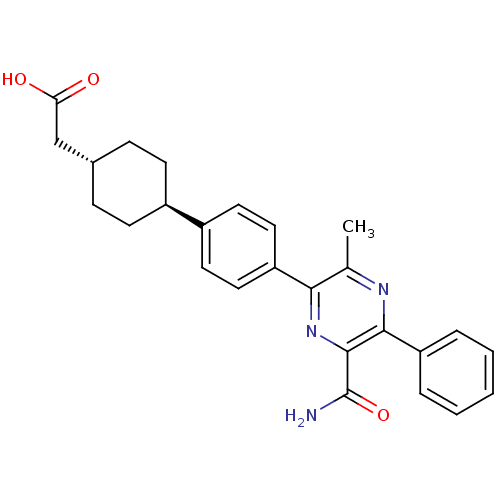
| ||