| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NAD-dependent protein deacetylase sirtuin-2 |
|---|
| Ligand | BDBM50400320 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_876635 (CHEMBL2182360) |
|---|
| IC50 | 24200±n/a nM |
|---|
| Citation |  Rotili, D; Tarantino, D; Nebbioso, A; Paolini, C; Huidobro, C; Lara, E; Mellini, P; Lenoci, A; Pezzi, R; Botta, G; Lahtela-Kakkonen, M; Poso, A; Steinkühler, C; Gallinari, P; De Maria, R; Fraga, M; Esteller, M; Altucci, L; Mai, A Discovery of salermide-related sirtuin inhibitors: binding mode studies and antiproliferative effects in cancer cells including cancer stem cells. J Med Chem55:10937-47 (2012) [PubMed] Article Rotili, D; Tarantino, D; Nebbioso, A; Paolini, C; Huidobro, C; Lara, E; Mellini, P; Lenoci, A; Pezzi, R; Botta, G; Lahtela-Kakkonen, M; Poso, A; Steinkühler, C; Gallinari, P; De Maria, R; Fraga, M; Esteller, M; Altucci, L; Mai, A Discovery of salermide-related sirtuin inhibitors: binding mode studies and antiproliferative effects in cancer cells including cancer stem cells. J Med Chem55:10937-47 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NAD-dependent protein deacetylase sirtuin-2 |
|---|
| Name: | NAD-dependent protein deacetylase sirtuin-2 |
|---|
| Synonyms: | NAD-Dependent Deacetylase Sirtuin-2 | NAD-dependent deacetylase sirtuin 2 | NAD-dependent deacetylase sirtuin 3 | NAD-dependent protein deacetylase sirtuin-2 (SIRT2) | SIR2-like | SIR2-like protein 2 | SIR2L | SIR2L2 | SIR2_HUMAN | SIRT2 | Sirtuin 2 (SIRT2) |
|---|
| Type: | Hydrolase |
|---|
| Mol. Mass.: | 43172.62 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 389 |
|---|
| Sequence: | MAEPDPSHPLETQAGKVQEAQDSDSDSEGGAAGGEADMDFLRNLFSQTLSLGSQKERLLD
ELTLEGVARYMQSERCRRVICLVGAGISTSAGIPDFRSPSTGLYDNLEKYHLPYPEAIFE
ISYFKKHPEPFFALAKELYPGQFKPTICHYFMRLLKDKGLLLRCYTQNIDTLERIAGLEQ
EDLVEAHGTFYTSHCVSASCRHEYPLSWMKEKIFSEVTPKCEDCQSLVKPDIVFFGESLP
ARFFSCMQSDFLKVDLLLVMGTSLQVQPFASLISKAPLSTPRLLINKEKAGQSDPFLGMI
MGLGGGMDFDSKKAYRDVAWLGECDQGCLALAELLGWKKELEDLVRREHASIDAQSGAGV
PNPSTSASPKKSPPPAKDEARTTEREKPQ
|
|
|
|---|
| BDBM50400320 |
|---|
| n/a |
|---|
| Name | BDBM50400320 |
|---|
| Synonyms: | CHEMBL2181509 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H23NO3S |
|---|
| Mol. Mass. | 429.531 |
|---|
| SMILES | CC(CS(=O)(=O)c1ccccc1\N=C\c1c(O)ccc2ccccc12)c1ccccc1 |
|---|
| Structure | 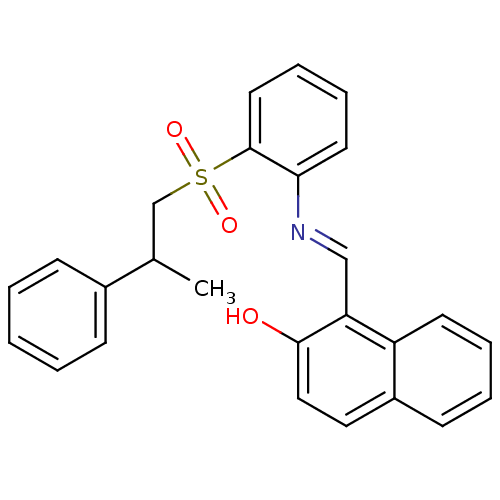
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Rotili, D; Tarantino, D; Nebbioso, A; Paolini, C; Huidobro, C; Lara, E; Mellini, P; Lenoci, A; Pezzi, R; Botta, G; Lahtela-Kakkonen, M; Poso, A; Steinkühler, C; Gallinari, P; De Maria, R; Fraga, M; Esteller, M; Altucci, L; Mai, A Discovery of salermide-related sirtuin inhibitors: binding mode studies and antiproliferative effects in cancer cells including cancer stem cells. J Med Chem55:10937-47 (2012) [PubMed] Article
Rotili, D; Tarantino, D; Nebbioso, A; Paolini, C; Huidobro, C; Lara, E; Mellini, P; Lenoci, A; Pezzi, R; Botta, G; Lahtela-Kakkonen, M; Poso, A; Steinkühler, C; Gallinari, P; De Maria, R; Fraga, M; Esteller, M; Altucci, L; Mai, A Discovery of salermide-related sirtuin inhibitors: binding mode studies and antiproliferative effects in cancer cells including cancer stem cells. J Med Chem55:10937-47 (2012) [PubMed] Article