| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone-lysine N-methyltransferase, H3 lysine-79 specific |
|---|
| Ligand | BDBM50400778 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_879145 (CHEMBL2209116) |
|---|
| IC50 | >100000±n/a nM |
|---|
| Citation |  Verma, SK; Tian, X; LaFrance, LV; Duquenne, C; Suarez, DP; Newlander, KA; Romeril, SP; Burgess, JL; Grant, SW; Brackley, JA; Graves, AP; Scherzer, DA; Shu, A; Thompson, C; Ott, HM; Aller, GS; Machutta, CA; Diaz, E; Jiang, Y; Johnson, NW; Knight, SD; Kruger, RG; McCabe, MT; Dhanak, D; Tummino, PJ; Creasy, CL; Miller, WH Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Med Chem Lett3:1091-1096 (2012) [PubMed] Article Verma, SK; Tian, X; LaFrance, LV; Duquenne, C; Suarez, DP; Newlander, KA; Romeril, SP; Burgess, JL; Grant, SW; Brackley, JA; Graves, AP; Scherzer, DA; Shu, A; Thompson, C; Ott, HM; Aller, GS; Machutta, CA; Diaz, E; Jiang, Y; Johnson, NW; Knight, SD; Kruger, RG; McCabe, MT; Dhanak, D; Tummino, PJ; Creasy, CL; Miller, WH Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Med Chem Lett3:1091-1096 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone-lysine N-methyltransferase, H3 lysine-79 specific |
|---|
| Name: | Histone-lysine N-methyltransferase, H3 lysine-79 specific |
|---|
| Synonyms: | 2.1.1.43 | DOT1-like protein | DOT1-like protein (Dot1L) | DOT1L | DOT1L_HUMAN | H3-K79-HMTase | Histone H3-K79 methyltransferase | Histone H3-K79 methyltransferase (DOT1L) | Histone Methyltransferase DOT1L | Histone-lysine N-methyltransferase, H3 lysine-79 specific (DOT1L) | KIAA1814 | KMT4 | Lysine N-methyltransferase 4 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 184911.91 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q8TEK3 |
|---|
| Residue: | 1537 |
|---|
| Sequence: | MGEKLELRLKSPVGAEPAVYPWPLPVYDKHHDAAHEIIETIRWVCEEIPDLKLAMENYVL
IDYDTKSFESMQRLCDKYNRAIDSIHQLWKGTTQPMKLNTRPSTGLLRHILQQVYNHSVT
DPEKLNNYEPFSPEVYGETSFDLVAQMIDEIKMTDDDLFVDLGSGVGQVVLQVAAATNCK
HHYGVEKADIPAKYAETMDREFRKWMKWYGKKHAEYTLERGDFLSEEWRERIANTSVIFV
NNFAFGPEVDHQLKERFANMKEGGRIVSSKPFAPLNFRINSRNLSDIGTIMRVVELSPLK
GSVSWTGKPVSYYLHTIDRTILENYFSSLKNPKLREEQEAARRRQQRESKSNAATPTKGP
EGKVAGPADAPMDSGAEEEKAGAATVKKPSPSKARKKKLNKKGRKMAGRKRGRPKKMNTA
NPERKPKKNQTALDALHAQTVSQTAASSPQDAYRSPHSPFYQLPPSVQRHSPNPLLVAPT
PPALQKLLESFKIQYLQFLAYTKTPQYKASLQELLGQEKEKNAQLLGAAQQLLSHCQAQK
EEIRRLFQQKLDELGVKALTYNDLIQAQKEISAHNQQLREQSEQLEQDNRALRGQSLQLL
KARCEELQLDWATLSLEKLLKEKQALKSQISEKQRHCLELQISIVELEKSQRQQELLQLK
SCVPPDDALSLHLRGKGALGRELEPDASRLHLELDCTKFSLPHLSSMSPELSMNGQAAGY
ELCGVLSRPSSKQNTPQYLASPLDQEVVPCTPSHVGRPRLEKLSGLAAPDYTRLSPAKIV
LRRHLSQDHTVPGRPAASELHSRAEHTKENGLPYQSPSVPGSMKLSPQDPRPLSPGALQL
AGEKSSEKGLRERAYGSSGELITSLPISIPLSTVQPNKLPVSIPLASVVLPSRAERARST
PSPVLQPRDPSSTLEKQIGANAHGAGSRSLALAPAGFSYAGSVAISGALAGSPASLTPGA
EPATLDESSSSGSLFATVGSRSSTPQHPLLLAQPRNSLPASPAHQLSSSPRLGGAAQGPL
PEASKGDLPSDSGFSDPESEAKRRIVFTITTGAGSAKQSPSSKHSPLTASARGDCVPSHG
QDSRRRGRRKRASAGTPSLSAGVSPKRRALPSVAGLFTQPSGSPLNLNSMVSNINQPLEI
TAISSPETSLKSSPVPYQDHDQPPVLKKERPLSQTNGAHYSPLTSDEEPGSEDEPSSARI
ERKIATISLESKSPPKTLENGGGLAGRKPAPAGEPVNSSKWKSTFSPISDIGLAKSADSP
LQASSALSQNSLFTFRPALEEPSADAKLAAHPRKGFPGSLSGADGLSPGTNPANGCTFGG
GLAADLSLHSFSDGASLPHKGPEAAGLSSPLSFPSQRGKEGSDANPFLSKRQLDGLAGLK
GEGSRGKEAGEGGLPLCGPTDKTPLLSGKAAKARDREVDLKNGHNLFISAAAVPPGSLLS
GPGLAPAASSAGGAASSAQTHRSFLGPFPPGPQFALGPMSLQANLGSVAGSSVLQSLFSS
VPAAAGLVHVSSAATRLTNSHAMGSFSGVAGGTVGGN
|
|
|
|---|
| BDBM50400778 |
|---|
| n/a |
|---|
| Name | BDBM50400778 |
|---|
| Synonyms: | CHEMBL2204995 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C31H39N7O2 |
|---|
| Mol. Mass. | 541.6871 |
|---|
| SMILES | CCCc1cc(C)[nH]c(=O)c1CNC(=O)c1cc(cc2n(ncc12)C(C)C)-c1ccnc(c1)N1CCN(C)CC1 |
|---|
| Structure | 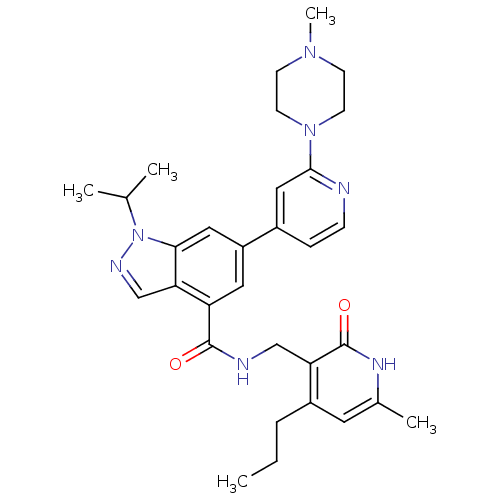
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Verma, SK; Tian, X; LaFrance, LV; Duquenne, C; Suarez, DP; Newlander, KA; Romeril, SP; Burgess, JL; Grant, SW; Brackley, JA; Graves, AP; Scherzer, DA; Shu, A; Thompson, C; Ott, HM; Aller, GS; Machutta, CA; Diaz, E; Jiang, Y; Johnson, NW; Knight, SD; Kruger, RG; McCabe, MT; Dhanak, D; Tummino, PJ; Creasy, CL; Miller, WH Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Med Chem Lett3:1091-1096 (2012) [PubMed] Article
Verma, SK; Tian, X; LaFrance, LV; Duquenne, C; Suarez, DP; Newlander, KA; Romeril, SP; Burgess, JL; Grant, SW; Brackley, JA; Graves, AP; Scherzer, DA; Shu, A; Thompson, C; Ott, HM; Aller, GS; Machutta, CA; Diaz, E; Jiang, Y; Johnson, NW; Knight, SD; Kruger, RG; McCabe, MT; Dhanak, D; Tummino, PJ; Creasy, CL; Miller, WH Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Med Chem Lett3:1091-1096 (2012) [PubMed] Article