| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Malate dehydrogenase, mitochondrial |
|---|
| Ligand | BDBM50404727 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_123822 (CHEMBL733653) |
|---|
| IC50 | 478630.09±n/a nM |
|---|
| Citation |  Coats, EA; Shah, KJ; Milstein, SR; Genther, CS; Nene, DM; Roesener, J; Schmidt, J; Pleiss, M; Wagner, E; Baker, JK 4-hydroxyquinoline-3-carboxylic acids as inhibitors of cell respiration. 2. Quantitative structure-activity relationship of dehydrogenase enzyme and Ehrlich ascites tumor cell inhibitions. J Med Chem25:57-63 (1982) [PubMed] Coats, EA; Shah, KJ; Milstein, SR; Genther, CS; Nene, DM; Roesener, J; Schmidt, J; Pleiss, M; Wagner, E; Baker, JK 4-hydroxyquinoline-3-carboxylic acids as inhibitors of cell respiration. 2. Quantitative structure-activity relationship of dehydrogenase enzyme and Ehrlich ascites tumor cell inhibitions. J Med Chem25:57-63 (1982) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Malate dehydrogenase, mitochondrial |
|---|
| Name: | Malate dehydrogenase, mitochondrial |
|---|
| Synonyms: | MDH2 | MDHM_PIG | Malate dehydrogenase mitochondrial |
|---|
| Type: | Oxidoreductase; homodimer |
|---|
| Mol. Mass.: | 35493.48 |
|---|
| Organism: | Sus scrofa (pig) |
|---|
| Description: | Porcine heart mitochondria. |
|---|
| Residue: | 337 |
|---|
| Sequence: | MLSALARPAGAALRRSFSTSQNNAKVAVLGASGGIGQPLSLLLKNSPLVSRLTLYDIAHT
PGVAADLSHIETRATVKGYLGPEQLPDCLKGCDVVVIPAGVPRKPGMTRDDLFNTNATIV
ATLTAACAQHCPDAMICIISNPVNSTIPITAEVFKKHGVYNPNKIFGVTTLDIVRANAFV
AELKGLDPARVSVPVIGGHAGKTIIPLISQCTPKVDFPQDQLSTLTGRIQEAGTEVVKAK
AGAGSATLSMAYAGARFVFSLVDAMNGKEGVVECSFVKSQETDCPYFSTPLLLGKKGIEK
NLGIGKISPFEEKMIAEAIPELKASIKKGEEFVKNMK
|
|
|
|---|
| BDBM50404727 |
|---|
| n/a |
|---|
| Name | BDBM50404727 |
|---|
| Synonyms: | CHEMBL138298 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C12H12N2O3 |
|---|
| Mol. Mass. | 232.2353 |
|---|
| SMILES | CN(C)c1ccc2c(O)c(cnc2c1)C(O)=O |
|---|
| Structure | 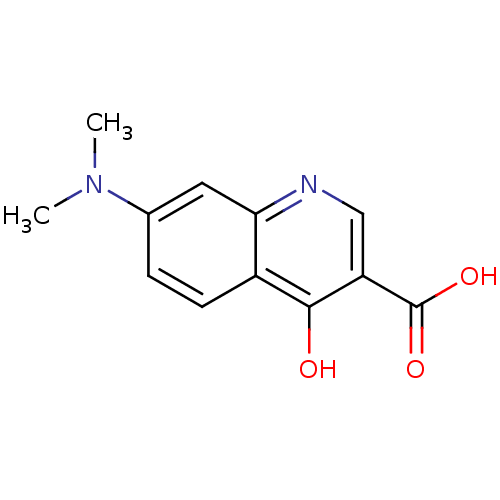
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Coats, EA; Shah, KJ; Milstein, SR; Genther, CS; Nene, DM; Roesener, J; Schmidt, J; Pleiss, M; Wagner, E; Baker, JK 4-hydroxyquinoline-3-carboxylic acids as inhibitors of cell respiration. 2. Quantitative structure-activity relationship of dehydrogenase enzyme and Ehrlich ascites tumor cell inhibitions. J Med Chem25:57-63 (1982) [PubMed]
Coats, EA; Shah, KJ; Milstein, SR; Genther, CS; Nene, DM; Roesener, J; Schmidt, J; Pleiss, M; Wagner, E; Baker, JK 4-hydroxyquinoline-3-carboxylic acids as inhibitors of cell respiration. 2. Quantitative structure-activity relationship of dehydrogenase enzyme and Ehrlich ascites tumor cell inhibitions. J Med Chem25:57-63 (1982) [PubMed]