| Reaction Details |
|---|
 | Report a problem with these data |
| Target | D(4) dopamine receptor |
|---|
| Ligand | BDBM50061349 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_307376 (CHEMBL875566) |
|---|
| Ki | 0.309±n/a nM |
|---|
| Citation |  Hansch, C; Verma, RP; Kurup, A; Mekapati, SB The role of QSAR in dopamine interactions. Bioorg Med Chem Lett15:2149-57 (2005) [PubMed] Article Hansch, C; Verma, RP; Kurup, A; Mekapati, SB The role of QSAR in dopamine interactions. Bioorg Med Chem Lett15:2149-57 (2005) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| D(4) dopamine receptor |
|---|
| Name: | D(4) dopamine receptor |
|---|
| Synonyms: | D(2C) dopamine receptor | DOPAMINE D4 | DOPAMINE D4.2 | DOPAMINE D4.4 | DRD4 | DRD4_HUMAN | Dopamine D4 receptor |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48373.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P21917 |
|---|
| Residue: | 419 |
|---|
| Sequence: | MGNRSTADADGLLAGRGPAAGASAGASAGLAGQGAAALVGGVLLIGAVLAGNSLVCVSVA
TERALQTPTNSFIVSLAAADLLLALLVLPLFVYSEVQGGAWLLSPRLCDALMAMDVMLCT
ASIFNLCAISVDRFVAVAVPLRYNRQGGSRRQLLLIGATWLLSAAVAAPVLCGLNDVRGR
DPAVCRLEDRDYVVYSSVCSFFLPCPLMLLLYWATFRGLQRWEVARRAKLHGRAPRRPSG
PGPPSPTPPAPRLPQDPCGPDCAPPAPGLPRGPCGPDCAPAAPSLPQDPCGPDCAPPAPG
LPPDPCGSNCAPPDAVRAAALPPQTPPQTRRRRRAKITGRERKAMRVLPVVVGAFLLCWT
PFFVVHITQALCPACSVPPRLVSAVTWLGYVNSALNPVIYTVFNAEFRNVFRKALRACC
|
|
|
|---|
| BDBM50061349 |
|---|
| n/a |
|---|
| Name | BDBM50061349 |
|---|
| Synonyms: | CHEMBL129534 | [2-(3-Chloro-phenoxy)-ethyl]-(3-phenoxy-propyl)-amine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H20ClNO2 |
|---|
| Mol. Mass. | 305.799 |
|---|
| SMILES | Clc1cccc(OCCNCCCOc2ccccc2)c1 |
|---|
| Structure | 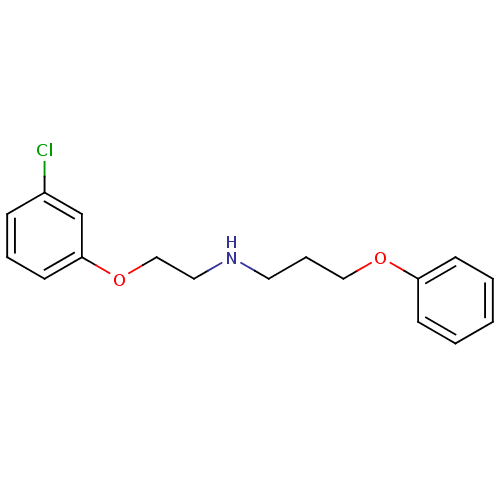
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hansch, C; Verma, RP; Kurup, A; Mekapati, SB The role of QSAR in dopamine interactions. Bioorg Med Chem Lett15:2149-57 (2005) [PubMed] Article
Hansch, C; Verma, RP; Kurup, A; Mekapati, SB The role of QSAR in dopamine interactions. Bioorg Med Chem Lett15:2149-57 (2005) [PubMed] Article