| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Muscarinic acetylcholine receptor M4 |
|---|
| Ligand | BDBM50241132 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_596258 (CHEMBL1048034) |
|---|
| Ki | 0.141±n/a nM |
|---|
| Citation |  Scapecchi, S; Nesi, M; Matucci, R; Bellucci, C; Buccioni, M; Dei, S; Guandalini, L; Manetti, D; Martelli, C; Martini, E; Marucci, G; Orlandi, F; Romanelli, MN; Teodori, E; Cirilli, R Synthesis, affinity profile and functional activity of potent chiral muscarinic antagonists with a pyrrolidinylfuran structure. J Med Chem53:201-7 (2010) [PubMed] Article Scapecchi, S; Nesi, M; Matucci, R; Bellucci, C; Buccioni, M; Dei, S; Guandalini, L; Manetti, D; Martelli, C; Martini, E; Marucci, G; Orlandi, F; Romanelli, MN; Teodori, E; Cirilli, R Synthesis, affinity profile and functional activity of potent chiral muscarinic antagonists with a pyrrolidinylfuran structure. J Med Chem53:201-7 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Muscarinic acetylcholine receptor M4 |
|---|
| Name: | Muscarinic acetylcholine receptor M4 |
|---|
| Synonyms: | ACM4_HUMAN | CHRM4 | Cholinergic, muscarinic M4 | Muscarinic acetylcholine receptor | Muscarinic acetylcholine receptor M2 and M4 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 53079.31 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Cholinergic, muscarinic M4 CHRM4 HUMAN M3::P08173 |
|---|
| Residue: | 479 |
|---|
| Sequence: | MANFTPVNGSSGNQSVRLVTSSSHNRYETVEMVFIATVTGSLSLVTVVGNILVMLSIKVN
RQLQTVNNYFLFSLACADLIIGAFSMNLYTVYIIKGYWPLGAVVCDLWLALDYVVSNASV
MNLLIISFDRYFCVTKPLTYPARRTTKMAGLMIAAAWVLSFVLWAPAILFWQFVVGKRTV
PDNQCFIQFLSNPAVTFGTAIAAFYLPVVIMTVLYIHISLASRSRVHKHRPEGPKEKKAK
TLAFLKSPLMKQSVKKPPPGEAAREELRNGKLEEAPPPALPPPPRPVADKDTSNESSSGS
ATQNTKERPATELSTTEATTPAMPAPPLQPRALNPASRWSKIQIVTKQTGNECVTAIEIV
PATPAGMRPAANVARKFASIARNQVRKKRQMAARERKVTRTIFAILLAFILTWTPYNVMV
LVNTFCQSCIPDTVWSIGYWLCYVNSTINPACYALCNATFKKTFRHLLLCQYRNIGTAR
|
|
|
|---|
| BDBM50241132 |
|---|
| n/a |
|---|
| Name | BDBM50241132 |
|---|
| Synonyms: | 3-Hydroxy-2-phenyl-propionic acid 9-methyl-3-oxa-9-aza-tricyclo[3.3.1.0*2,4*]non-7-yl ester | 7-(3-Hydroxy-2-phenyl-propionyloxy)-9,9-dimethyl-3-oxa-9-azonia-tricyclo[3.3.1.0*2,4*]nonane | CHEMBL376897 | METHSCOPOLAMINE | Methyl scopolamine | N-METHYLSCOPOLAMINE |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H24NO4 |
|---|
| Mol. Mass. | 318.3869 |
|---|
| SMILES | C[N+]1(C)[C@H]2C[C@@H](C[C@@H]1[C@H]1O[C@@H]21)OC(=O)[C@H](CO)c1ccccc1 |r,TLB:9:8:4.5.6:1,9:10:4.5.6:1,THB:11:5:1:8.10| |
|---|
| Structure | 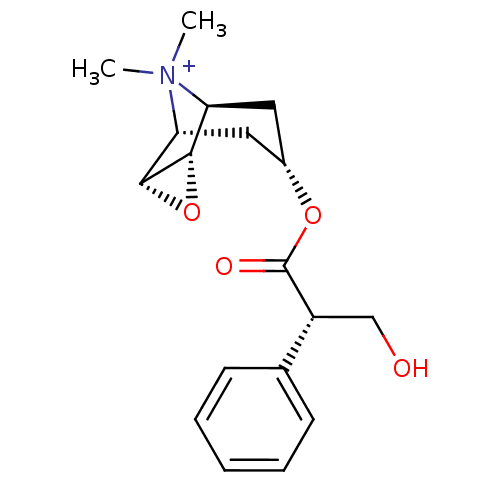
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Scapecchi, S; Nesi, M; Matucci, R; Bellucci, C; Buccioni, M; Dei, S; Guandalini, L; Manetti, D; Martelli, C; Martini, E; Marucci, G; Orlandi, F; Romanelli, MN; Teodori, E; Cirilli, R Synthesis, affinity profile and functional activity of potent chiral muscarinic antagonists with a pyrrolidinylfuran structure. J Med Chem53:201-7 (2010) [PubMed] Article
Scapecchi, S; Nesi, M; Matucci, R; Bellucci, C; Buccioni, M; Dei, S; Guandalini, L; Manetti, D; Martelli, C; Martini, E; Marucci, G; Orlandi, F; Romanelli, MN; Teodori, E; Cirilli, R Synthesis, affinity profile and functional activity of potent chiral muscarinic antagonists with a pyrrolidinylfuran structure. J Med Chem53:201-7 (2010) [PubMed] Article