

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Serine/threonine-protein kinase 3 | ||
| Ligand | BDBM50425862 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_933532 (CHEMBL2319694) | ||
| IC50 | 470±n/a nM | ||
| Citation |  Powell, NA; Hoffman, JK; Ciske, FL; Kohrt, JT; Baxi, SM; Peng, YW; Zhong, M; Catana, C; Ohren, J; Perrin, LA; Edmunds, JJ Optimization of highly selective 2,4-diaminopyrimidine-5-carboxamide inhibitors of Sky kinase. Bioorg Med Chem Lett23:1051-5 (2013) [PubMed] Article Powell, NA; Hoffman, JK; Ciske, FL; Kohrt, JT; Baxi, SM; Peng, YW; Zhong, M; Catana, C; Ohren, J; Perrin, LA; Edmunds, JJ Optimization of highly selective 2,4-diaminopyrimidine-5-carboxamide inhibitors of Sky kinase. Bioorg Med Chem Lett23:1051-5 (2013) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Serine/threonine-protein kinase 3 | |||
| Name: | Serine/threonine-protein kinase 3 | ||
| Synonyms: | KRS1 | MST-2 | MST2 | Mammalian STE20-like protein kinase 2 | STE20-Like Kinase MST2 | STK3 | STK3_HUMAN | Serine/threonine-protein kinase 3 | Serine/threonine-protein kinase 3 (MST2) | Serine/threonine-protein kinase 3/4 | Serine/threonine-protein kinase Krs-1 | Serine/threonine-protein kinase MST2 | ||
| Type: | Serine/threonine-protein kinase | ||
| Mol. Mass.: | 56284.87 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 491 | ||
| Sequence: |
| ||
| BDBM50425862 | |||
| n/a | |||
| Name | BDBM50425862 | ||
| Synonyms: | CHEMBL2312654 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H26Cl2N6O | ||
| Mol. Mass. | 461.387 | ||
| SMILES | CN(C)C1CCC(CC1)Nc1nc(Nc2cc(Cl)cc(Cl)c2)ncc1-c1cc(C)no1 |(9.04,-43.08,;10.39,-42.35,;11.7,-43.16,;10.44,-40.81,;9.13,-40,;9.18,-38.46,;10.53,-37.74,;11.85,-38.53,;11.8,-40.08,;10.57,-36.2,;9.81,-34.87,;8.27,-34.86,;7.51,-33.53,;5.96,-33.52,;5.2,-32.19,;5.97,-30.85,;5.21,-29.52,;5.98,-28.19,;3.67,-29.52,;2.89,-30.86,;1.35,-30.86,;3.66,-32.19,;8.27,-32.2,;9.8,-32.19,;10.58,-33.53,;12.11,-33.53,;12.6,-34.99,;14.14,-34.99,;15.06,-36.23,;14.61,-33.52,;13.35,-32.62,)| | ||
| Structure | 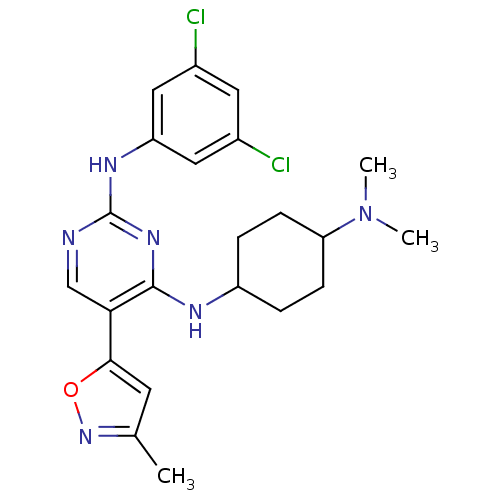
| ||