| Reaction Details |
|---|
 | Report a problem with these data |
| Target | D-amino-acid oxidase |
|---|
| Ligand | BDBM36181 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_938787 (CHEMBL2327931) |
|---|
| IC50 | 65700±n/a nM |
|---|
| Citation |  Katane, M; Osaka, N; Matsuda, S; Maeda, K; Kawata, T; Saitoh, Y; Sekine, M; Furuchi, T; Doi, I; Hirono, S; Homma, H Identification of novel D-amino acid oxidase inhibitors by in silico screening and their functional characterization in vitro. J Med Chem56:1894-907 (2013) [PubMed] Article Katane, M; Osaka, N; Matsuda, S; Maeda, K; Kawata, T; Saitoh, Y; Sekine, M; Furuchi, T; Doi, I; Hirono, S; Homma, H Identification of novel D-amino acid oxidase inhibitors by in silico screening and their functional characterization in vitro. J Med Chem56:1894-907 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| D-amino-acid oxidase |
|---|
| Name: | D-amino-acid oxidase |
|---|
| Synonyms: | D-amino-acid oxidase (DAAO) | DAAO | DAMOX | DAO | OXDA_HUMAN |
|---|
| Type: | Homodimer |
|---|
| Mol. Mass.: | 39476.06 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P14920 |
|---|
| Residue: | 347 |
|---|
| Sequence: | MRVVVIGAGVIGLSTALCIHERYHSVLQPLDIKVYADRFTPLTTTDVAAGLWQPYLSDPN
NPQEADWSQQTFDYLLSHVHSPNAENLGLFLISGYNLFHEAIPDPSWKDTVLGFRKLTPR
ELDMFPDYGYGWFHTSLILEGKNYLQWLTERLTERGVKFFQRKVESFEEVAREGADVIVN
CTGVWAGALQRDPLLQPGRGQIMKVDAPWMKHFILTHDPERGIYNSPYIIPGTQTVTLGG
IFQLGNWSELNNIQDHNTIWEGCCRLEPTLKNARIIGERTGFRPVRPQIRLEREQLRTGP
SNTEVIHNYGHGGYGLTIHWGCALEAAKLFGRILEEKKLSRMPPSHL
|
|
|
|---|
| BDBM36181 |
|---|
| n/a |
|---|
| Name | BDBM36181 |
|---|
| Synonyms: | SODIUM BENZOATE | benzoate | benzoic acid | benzoic acid-d5 |
|---|
| Type | Guest |
|---|
| Emp. Form. | C7H5O2 |
|---|
| Mol. Mass. | 121.1139 |
|---|
| SMILES | [O-]C(=O)c1ccccc1 |
|---|
| Structure | 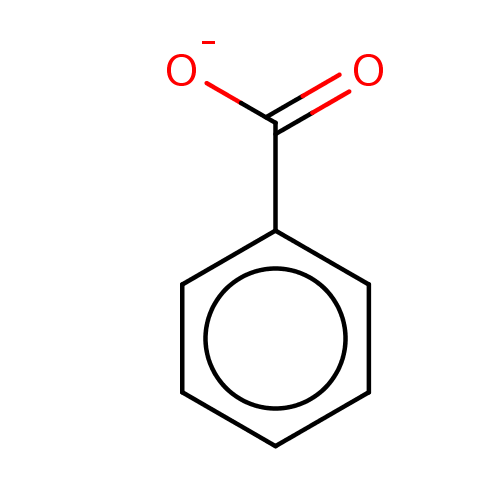
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Katane, M; Osaka, N; Matsuda, S; Maeda, K; Kawata, T; Saitoh, Y; Sekine, M; Furuchi, T; Doi, I; Hirono, S; Homma, H Identification of novel D-amino acid oxidase inhibitors by in silico screening and their functional characterization in vitro. J Med Chem56:1894-907 (2013) [PubMed] Article
Katane, M; Osaka, N; Matsuda, S; Maeda, K; Kawata, T; Saitoh, Y; Sekine, M; Furuchi, T; Doi, I; Hirono, S; Homma, H Identification of novel D-amino acid oxidase inhibitors by in silico screening and their functional characterization in vitro. J Med Chem56:1894-907 (2013) [PubMed] Article