

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Protein arginine N-methyltransferase 3 [N508S] | ||
| Ligand | BDBM50427798 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_941988 (CHEMBL2330282) | ||
| IC50 | >10000±n/a nM | ||
| Citation |  Liu, F; Li, F; Ma, A; Dobrovetsky, E; Dong, A; Gao, C; Korboukh, I; Liu, J; Smil, D; Brown, PJ; Frye, SV; Arrowsmith, CH; Schapira, M; Vedadi, M; Jin, J Exploiting an allosteric binding site of PRMT3 yields potent and selective inhibitors. J Med Chem56:2110-24 (2013) [PubMed] Article Liu, F; Li, F; Ma, A; Dobrovetsky, E; Dong, A; Gao, C; Korboukh, I; Liu, J; Smil, D; Brown, PJ; Frye, SV; Arrowsmith, CH; Schapira, M; Vedadi, M; Jin, J Exploiting an allosteric binding site of PRMT3 yields potent and selective inhibitors. J Med Chem56:2110-24 (2013) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Protein arginine N-methyltransferase 3 [N508S] | |||
| Name: | Protein arginine N-methyltransferase 3 [N508S] | ||
| Synonyms: | ANM3_HUMAN | HRMT1L3 | Heterogeneous nuclear ribonucleoprotein methyltransferase-like protein 3 | PRMT3 | Protein arginine N-methyltransferase 3 | Protein arginine N-methyltransferase 3 (PRMT3) | ||
| Type: | Protein | ||
| Mol. Mass.: | 59859.85 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | O60678[N508S] | ||
| Residue: | 531 | ||
| Sequence: |
| ||
| BDBM50427798 | |||
| n/a | |||
| Name | BDBM50427798 | ||
| Synonyms: | CHEMBL2325187 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C15H21N5OS | ||
| Mol. Mass. | 319.425 | ||
| SMILES | N[C@H]1CC[C@H](CCNC(=O)Nc2ccc3nnsc3c2)CC1 |r,wU:4.4,wD:1.0,(39.54,-53.05,;38.2,-53.82,;36.87,-53.05,;35.53,-53.81,;35.54,-55.35,;34.21,-56.12,;32.87,-55.35,;31.54,-56.13,;30.21,-55.36,;30.2,-53.82,;28.87,-56.13,;27.54,-55.36,;27.53,-53.81,;26.2,-53.05,;24.87,-53.82,;23.4,-53.34,;22.49,-54.6,;23.4,-55.84,;24.87,-55.36,;26.2,-56.13,;36.87,-56.12,;38.19,-55.36,)| | ||
| Structure | 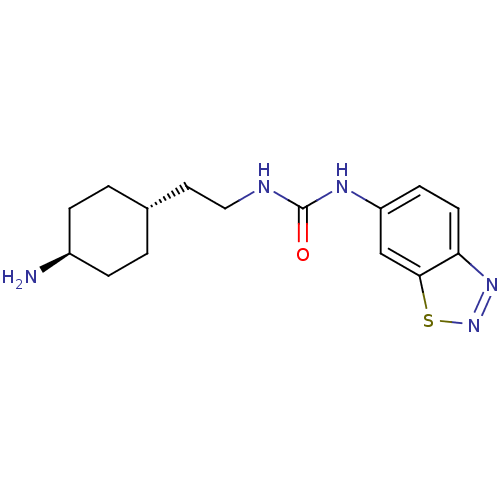
| ||