| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NAD-dependent histone deacetylase SIR2 |
|---|
| Ligand | BDBM29590 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_942450 (CHEMBL2345217) |
|---|
| IC50 | 60000±n/a nM |
|---|
| Citation |  Rambabu, D; Raja, G; Yogi Sreenivas, B; Seerapu, GP; Lalith Kumar, K; Deora, GS; Haldar, D; Rao, MV; Pal, M Spiro heterocycles as potential inhibitors of SIRT1: Pd/C-mediated synthesis of novel N-indolylmethyl spiroindoline-3,2'-quinazolines. Bioorg Med Chem Lett23:1351-7 (2013) [PubMed] Article Rambabu, D; Raja, G; Yogi Sreenivas, B; Seerapu, GP; Lalith Kumar, K; Deora, GS; Haldar, D; Rao, MV; Pal, M Spiro heterocycles as potential inhibitors of SIRT1: Pd/C-mediated synthesis of novel N-indolylmethyl spiroindoline-3,2'-quinazolines. Bioorg Med Chem Lett23:1351-7 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NAD-dependent histone deacetylase SIR2 |
|---|
| Name: | NAD-dependent histone deacetylase SIR2 |
|---|
| Synonyms: | MAR1 | SIR2 | SIR2_YEAST |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 63277.80 |
|---|
| Organism: | Saccharomyces cerevisiae |
|---|
| Description: | ChEMBL_942450 |
|---|
| Residue: | 562 |
|---|
| Sequence: | MTIPHMKYAVSKTSENKVSNTVSPTQDKDAIRKQPDDIINNDEPSHKKIKVAQPDSLRET
NTTDPLGHTKAALGEVASMELKPTNDMDPLAVSAASVVSMSNDVLKPETPKGPIIISKNP
SNGIFYGPSFTKRESLNARMFLKYYGAHKFLDTYLPEDLNSLYIYYLIKLLGFEVKDQAL
IGTINSIVHINSQERVQDLGSAISVTNVEDPLAKKQTVRLIKDLQRAINKVLCTRLRLSN
FFTIDHFIQKLHTARKILVLTGAGVSTSLGIPDFRSSEGFYSKIKHLGLDDPQDVFNYNI
FMHDPSVFYNIANMVLPPEKIYSPLHSFIKMLQMKGKLLRNYTQNIDNLESYAGISTDKL
VQCHGSFATATCVTCHWNLPGERIFNKIRNLELPLCPYCYKKRREYFPEGYNNKVGVAAS
QGSMSERPPYILNSYGVLKPDITFFGEALPNKFHKSIREDILECDLLICIGTSLKVAPVS
EIVNMVPSHVPQVLINRDPVKHAEFDLSLLGYCDDIAAMVAQKCGWTIPHKKWNDLKNKN
FKCQEKDKGVYVVTSDEHPKTL
|
|
|
|---|
| BDBM29590 |
|---|
| n/a |
|---|
| Name | BDBM29590 |
|---|
| Synonyms: | 1,2-Dihydro-3H-naphtho[2,1-b]pyran-3-one | CHEMBL86537 | splitomicin |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H10O2 |
|---|
| Mol. Mass. | 198.2173 |
|---|
| SMILES | O=C1CCc2c(O1)ccc1ccccc21 |
|---|
| Structure | 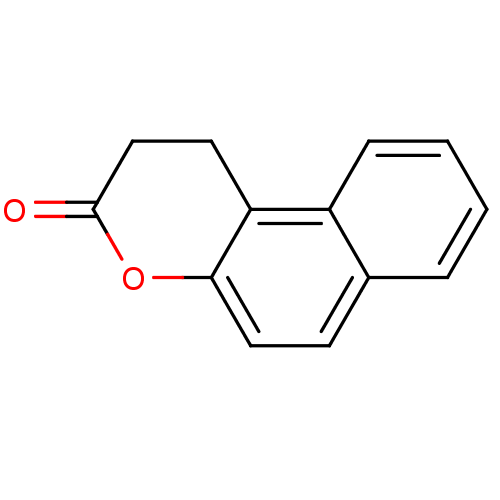
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Rambabu, D; Raja, G; Yogi Sreenivas, B; Seerapu, GP; Lalith Kumar, K; Deora, GS; Haldar, D; Rao, MV; Pal, M Spiro heterocycles as potential inhibitors of SIRT1: Pd/C-mediated synthesis of novel N-indolylmethyl spiroindoline-3,2'-quinazolines. Bioorg Med Chem Lett23:1351-7 (2013) [PubMed] Article
Rambabu, D; Raja, G; Yogi Sreenivas, B; Seerapu, GP; Lalith Kumar, K; Deora, GS; Haldar, D; Rao, MV; Pal, M Spiro heterocycles as potential inhibitors of SIRT1: Pd/C-mediated synthesis of novel N-indolylmethyl spiroindoline-3,2'-quinazolines. Bioorg Med Chem Lett23:1351-7 (2013) [PubMed] Article