| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Poly [ADP-ribose] polymerase 2 |
|---|
| Ligand | BDBM50438604 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_975899 (CHEMBL2415548) |
|---|
| IC50 | 480±n/a nM |
|---|
| Citation |  Woon, EC; Sunderland, PT; Paine, HA; Lloyd, MD; Thompson, AS; Threadgill, MD One-pot tandem Hurtley-retro-Claisen-cyclisation reactions in the synthesis of 3-substituted analogues of 5-aminoisoquinolin-1-one (5-AIQ), a water-soluble inhibitor of PARPs. Bioorg Med Chem21:5218-27 (2013) [PubMed] Article Woon, EC; Sunderland, PT; Paine, HA; Lloyd, MD; Thompson, AS; Threadgill, MD One-pot tandem Hurtley-retro-Claisen-cyclisation reactions in the synthesis of 3-substituted analogues of 5-aminoisoquinolin-1-one (5-AIQ), a water-soluble inhibitor of PARPs. Bioorg Med Chem21:5218-27 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Poly [ADP-ribose] polymerase 2 |
|---|
| Name: | Poly [ADP-ribose] polymerase 2 |
|---|
| Synonyms: | 2.4.2.- | 2.4.2.30 | ADP-ribosyltransferase diphtheria toxin-like 2 | ADPRT-2 | ARTD2 | Adprt2 | Adprtl2 | Aspartl2 | DNA ADP-ribosyltransferase PARP2 | NAD(+) ADP-ribosyltransferase 2 | PARP-2 | PARP2_MOUSE | Parp2 | Poly (ADP-ribose) Polymerase-2 (PARP-2) | Poly [ADP-ribose] polymerase 2 (Parp2) | Poly [ADP-ribose] polymerase-2 | Poly[ADP-ribose] synthase 2 | Poly[ADP-ribose] synthetase 2 | Protein poly-ADP-ribosyltransferase PARP2 | mPARP-2 | pADPRT-2 | poly-ADP-ribose polymerase 2 (PARP2) |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 63409.97 |
|---|
| Organism: | Mus musculus (Mouse) |
|---|
| Description: | Alexis Biochemicals, Cat. no. ALX-201-064-C020 |
|---|
| Residue: | 559 |
|---|
| Sequence: | MAPRRQRSGSGRRVLNEAKKVDNGNKATEDDSPPGKKMRTCQRKGPMAGGKDADRTKDNR
DSVKTLLLKGKAPVDPECAAKLGKAHVYCEGDDVYDVMLNQTNLQFNNNKYYLIQLLEDD
AQRNFSVWMRWGRVGKTGQHSLVTCSGDLNKAKEIFQKKFLDKTKNNWEDRENFEKVPGK
YDMLQMDYAASTQDESKTKEEETLKPESQLDLRVQELLKLICNVQTMEEMMIEMKYDTKR
APLGKLTVAQIKAGYQSLKKIEDCIRAGQHGRALVEACNEFYTRIPHDFGLSIPPVIRTE
KELSDKVKLLEALGDIEIALKLVKSERQGLEHPLDQHYRNLHCALRPLDHESNEFKVISQ
YLQSTHAPTHKDYTMTLLDVFEVEKEGEKEAFREDLPNRMLLWHGSRLSNWVGILSHGLR
VAPPEAPITGYMFGKGIYFADMSSKSANYCFASRLKNTGLLLLSEVALGQCNELLEANPK
AQGLLRGKHSTKGMGKMAPSPAHFITLNGSTVPLGPASDTGILNPEGYTLNYNEFIVYSP
NQVRMRYLLKIQFNFLQLW
|
|
|
|---|
| BDBM50438604 |
|---|
| n/a |
|---|
| Name | BDBM50438604 |
|---|
| Synonyms: | CHEMBL2414043 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H12N2O |
|---|
| Mol. Mass. | 236.2686 |
|---|
| SMILES | Nc1cccc2c1cc([nH]c2=O)-c1ccccc1 |
|---|
| Structure | 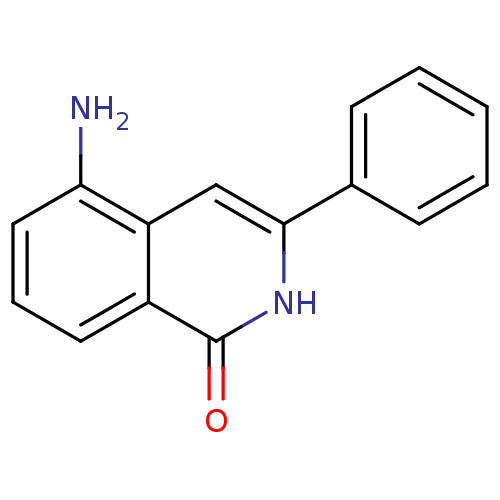
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Woon, EC; Sunderland, PT; Paine, HA; Lloyd, MD; Thompson, AS; Threadgill, MD One-pot tandem Hurtley-retro-Claisen-cyclisation reactions in the synthesis of 3-substituted analogues of 5-aminoisoquinolin-1-one (5-AIQ), a water-soluble inhibitor of PARPs. Bioorg Med Chem21:5218-27 (2013) [PubMed] Article
Woon, EC; Sunderland, PT; Paine, HA; Lloyd, MD; Thompson, AS; Threadgill, MD One-pot tandem Hurtley-retro-Claisen-cyclisation reactions in the synthesis of 3-substituted analogues of 5-aminoisoquinolin-1-one (5-AIQ), a water-soluble inhibitor of PARPs. Bioorg Med Chem21:5218-27 (2013) [PubMed] Article