| Reaction Details |
|---|
 | Report a problem with these data |
| Target | DNA-(apurinic or apyrimidinic site) endonuclease |
|---|
| Ligand | BDBM34905 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_978866 (CHEMBL2423657) |
|---|
| IC50 | >200000±n/a nM |
|---|
| Citation |  Raoof, A; Depledge, P; Hamilton, NM; Hamilton, NS; Hitchin, JR; Hopkins, GV; Jordan, AM; Maguire, LA; McGonagle, AE; Mould, DP; Rushbrooke, M; Small, HF; Smith, KM; Thomson, GJ; Turlais, F; Waddell, ID; Waszkowycz, B; Watson, AJ; Ogilvie, DJ Toxoflavins and deazaflavins as the first reported selective small molecule inhibitors of tyrosyl-DNA phosphodiesterase II. J Med Chem56:6352-70 (2013) [PubMed] Article Raoof, A; Depledge, P; Hamilton, NM; Hamilton, NS; Hitchin, JR; Hopkins, GV; Jordan, AM; Maguire, LA; McGonagle, AE; Mould, DP; Rushbrooke, M; Small, HF; Smith, KM; Thomson, GJ; Turlais, F; Waddell, ID; Waszkowycz, B; Watson, AJ; Ogilvie, DJ Toxoflavins and deazaflavins as the first reported selective small molecule inhibitors of tyrosyl-DNA phosphodiesterase II. J Med Chem56:6352-70 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| DNA-(apurinic or apyrimidinic site) endonuclease |
|---|
| Name: | DNA-(apurinic or apyrimidinic site) endonuclease |
|---|
| Synonyms: | APE | APE1 | APEX | APEX1 | APEX1_HUMAN | APX | Apurinic-apyrimidinic endonuclease 1 (APE-1) | Apurinic/apyrimidinic endonuclease 1 (APE1) | DNA-(apurinic or apyrimidinic site) lyase | HAP1 | REF1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 35560.12 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P27695 |
|---|
| Residue: | 318 |
|---|
| Sequence: | MPKRGKKGAVAEDGDELRTEPEAKKSKTAAKKNDKEAAGEGPALYEDPPDQKTSPSGKPA
TLKICSWNVDGLRAWIKKKGLDWVKEEAPDILCLQETKCSENKLPAELQELPGLSHQYWS
APSDKEGYSGVGLLSRQCPLKVSYGIGDEEHDQEGRVIVAEFDSFVLVTAYVPNAGRGLV
RLEYRQRWDEAFRKFLKGLASRKPLVLCGDLNVAHEEIDLRNPKGNKKNAGFTPQERQGF
GELLQAVPLADSFRHLYPNTPYAYTFWTYMMNARSKNVGWRLDYFLLSHSLLPALCDSKI
RSKALGSDHCPITLYLAL
|
|
|
|---|
| BDBM34905 |
|---|
| n/a |
|---|
| Name | BDBM34905 |
|---|
| Synonyms: | 1,6-dimethyl-3-propyl-pyrimido[5,4-e][1,2,4]triazine-5,7-dione | 1,6-dimethyl-3-propyl-pyrimido[5,4-e][1,2,4]triazine-5,7-quinone | 1,6-dimethyl-3-propylpyrimido[5,4-e][1,2,4]triazine-5,7-dione | MLS000718932 | SMR000291200 | US9073941, 617 | cid_3164059 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H13N5O2 |
|---|
| Mol. Mass. | 235.2425 |
|---|
| SMILES | CCCc1nn(C)c2nc(=O)n(C)c(=O)c2n1 |
|---|
| Structure | 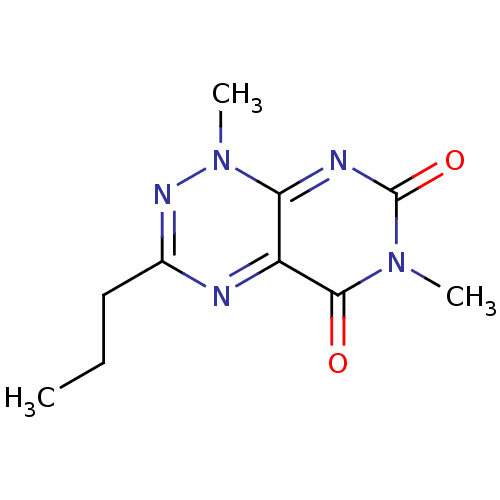
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Raoof, A; Depledge, P; Hamilton, NM; Hamilton, NS; Hitchin, JR; Hopkins, GV; Jordan, AM; Maguire, LA; McGonagle, AE; Mould, DP; Rushbrooke, M; Small, HF; Smith, KM; Thomson, GJ; Turlais, F; Waddell, ID; Waszkowycz, B; Watson, AJ; Ogilvie, DJ Toxoflavins and deazaflavins as the first reported selective small molecule inhibitors of tyrosyl-DNA phosphodiesterase II. J Med Chem56:6352-70 (2013) [PubMed] Article
Raoof, A; Depledge, P; Hamilton, NM; Hamilton, NS; Hitchin, JR; Hopkins, GV; Jordan, AM; Maguire, LA; McGonagle, AE; Mould, DP; Rushbrooke, M; Small, HF; Smith, KM; Thomson, GJ; Turlais, F; Waddell, ID; Waszkowycz, B; Watson, AJ; Ogilvie, DJ Toxoflavins and deazaflavins as the first reported selective small molecule inhibitors of tyrosyl-DNA phosphodiesterase II. J Med Chem56:6352-70 (2013) [PubMed] Article