

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Neuronal acetylcholine receptor subunit alpha-3/beta-4 | ||
| Ligand | BDBM50446488 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1287913 (CHEMBL3112332) | ||
| Ki | 78400±n/a nM | ||
| Citation |  Wu, J; Zhang, Y; Maida, LE; Santos, RG; Welmaker, GS; LaVoi, TM; Nefzi, A; Yu, Y; Houghten, RA; Toll, L; Giulianotti, MA Scaffold ranking and positional scanning utilized in the discovery of nAChR-selective compounds suitable for optimization studies. J Med Chem56:10103-17 (2013) [PubMed] Article Wu, J; Zhang, Y; Maida, LE; Santos, RG; Welmaker, GS; LaVoi, TM; Nefzi, A; Yu, Y; Houghten, RA; Toll, L; Giulianotti, MA Scaffold ranking and positional scanning utilized in the discovery of nAChR-selective compounds suitable for optimization studies. J Med Chem56:10103-17 (2013) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Neuronal acetylcholine receptor subunit alpha-3/beta-4 | |||
| Name: | Neuronal acetylcholine receptor subunit alpha-3/beta-4 | ||
| Synonyms: | Neuronal acetylcholine receptor Alpha-3/Beta-4 | Neuronal acetylcholine receptor protein alpha-3/beta-4 subunit | Neuronal acetylcholine receptor; alpha3/beta4 | nAChR subtypes alpha3 beta4 | ||
| Type: | Protein | ||
| Mol. Mass.: | n/a | ||
| Description: | n/a | ||
| Components: | This complex has 2 components. | ||
| Component 1 | |||
| Name: | Neuronal acetylcholine receptor subunit alpha-3 | ||
| Synonyms: | ACHA3_RAT | Acra3 | Chrna3 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 56995.52 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | P04757 | ||
| Residue: | 499 | ||
| Sequence: |
| ||
| Component 2 | |||
| Name: | Neuronal acetylcholine receptor subunit beta-4 | ||
| Synonyms: | ACHB4_RAT | Acrb4 | Chrnb4 | N-alpha 2 | Neuronal acetylcholine receptor non-alpha-2 chain | Neuronal acetylcholine receptor protein beta-4 subunit | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 55863.89 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | P12392 | ||
| Residue: | 495 | ||
| Sequence: |
| ||
| BDBM50446488 | |||
| n/a | |||
| Name | BDBM50446488 | ||
| Synonyms: | CHEMBL3110036 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C34H64N6 | ||
| Mol. Mass. | 556.9122 | ||
| SMILES | CC(C)CCCN1[C@@H](CCCCN2C[C@@H](CC3CCCCC3)N(CC3CCC(CC3)C(C)(C)C)C2=N)CN=C1N |r,wU:7.7,wD:14.14,c:40,(29.06,-42.63,;27.52,-42.56,;26.81,-41.19,;26.69,-43.85,;27.4,-45.22,;26.56,-46.52,;27.27,-47.88,;28.74,-48.34,;30.08,-47.59,;31.4,-48.38,;32.75,-47.62,;34.08,-48.4,;35.41,-47.65,;36.89,-48.1,;37.77,-46.84,;39.31,-46.82,;40.06,-45.47,;41.6,-45.46,;42.35,-44.12,;41.57,-42.8,;40.03,-42.82,;39.27,-44.16,;36.85,-45.61,;37.52,-44.22,;36.7,-42.92,;37.42,-41.57,;36.6,-40.27,;35.06,-40.33,;34.35,-41.69,;35.17,-43,;34.24,-39.03,;34.96,-37.67,;32.7,-39.09,;33.46,-37.68,;35.39,-46.11,;34.13,-45.22,;28.75,-49.88,;27.29,-50.38,;26.38,-49.14,;24.84,-49.15,)| | ||
| Structure | 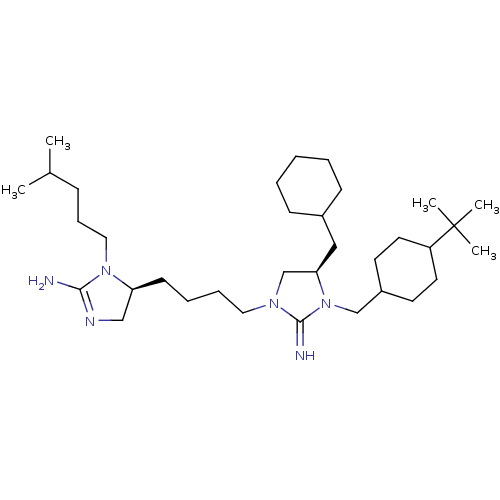
| ||