| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytochrome P450 1A2 |
|---|
| Ligand | BDBM520 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1291569 (CHEMBL3118925) |
|---|
| IC50 | >25000±n/a nM |
|---|
| Citation |  Liu, H; Xu, L; Hui, H; Vivian, R; Callebaut, C; Murray, BP; Hong, A; Lee, MS; Tsai, LK; Chau, JK; Stray, KM; Cannizzaro, C; Choi, YC; Rhodes, GR; Desai, MC Structure-activity relationships of diamine inhibitors of cytochrome P450 (CYP) 3A as novel pharmacoenhancers, part I: core region. Bioorg Med Chem Lett24:989-94 (2014) [PubMed] Article Liu, H; Xu, L; Hui, H; Vivian, R; Callebaut, C; Murray, BP; Hong, A; Lee, MS; Tsai, LK; Chau, JK; Stray, KM; Cannizzaro, C; Choi, YC; Rhodes, GR; Desai, MC Structure-activity relationships of diamine inhibitors of cytochrome P450 (CYP) 3A as novel pharmacoenhancers, part I: core region. Bioorg Med Chem Lett24:989-94 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytochrome P450 1A2 |
|---|
| Name: | Cytochrome P450 1A2 |
|---|
| Synonyms: | CP1A2_HUMAN | CYP1A2 | CYPIA2 | Cholesterol 25-hydroxylase | Cytochrome P(3)450 | Cytochrome P450 1A | Cytochrome P450 1A2 (CYP1A2) | Cytochrome P450 4 | Cytochrome P450-P3 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 58423.38 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P05177 |
|---|
| Residue: | 516 |
|---|
| Sequence: | MALSQSVPFSATELLLASAIFCLVFWVLKGLRPRVPKGLKSPPEPWGWPLLGHVLTLGKN
PHLALSRMSQRYGDVLQIRIGSTPVLVLSRLDTIRQALVRQGDDFKGRPDLYTSTLITDG
QSLTFSTDSGPVWAARRRLAQNALNTFSIASDPASSSSCYLEEHVSKEAKALISRLQELM
AGPGHFDPYNQVVVSVANVIGAMCFGQHFPESSDEMLSLVKNTHEFVETASSGNPLDFFP
ILRYLPNPALQRFKAFNQRFLWFLQKTVQEHYQDFDKNSVRDITGALFKHSKKGPRASGN
LIPQEKIVNLVNDIFGAGFDTVTTAISWSLMYLVTKPEIQRKIQKELDTVIGRERRPRLS
DRPQLPYLEAFILETFRHSSFLPFTIPHSTTRDTTLNGFYIPKKCCVFVNQWQVNHDPEL
WEDPSEFRPERFLTADGTAINKPLSEKMMLFGMGKRRCIGEVLAKWEIFLFLAILLQQLE
FSVPPGVKVDLTPIYGLTMKHARCEHVQARLRFSIN
|
|
|
|---|
| BDBM520 |
|---|
| n/a |
|---|
| Name | BDBM520 |
|---|
| Synonyms: | 1,3-thiazol-5-ylmethyl N-[(2S,3S,5S)-3-hydroxy-5-[(2S)-3-methyl-2-{[methyl({[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl})carbamoyl]amino}butanamido]-1,6-diphenylhexan-2-yl]carbamate | ABT-538 | CHEMBL163 | Norvir | RTV | Ritonavir |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C37H48N6O5S2 |
|---|
| Mol. Mass. | 720.944 |
|---|
| SMILES | CC(C)[C@H](NC(=O)N(C)Cc1csc(n1)C(C)C)C(=O)N[C@H](C[C@H](O)[C@H](Cc1ccccc1)NC(=O)OCc1cncs1)Cc1ccccc1 |
|---|
| Structure | 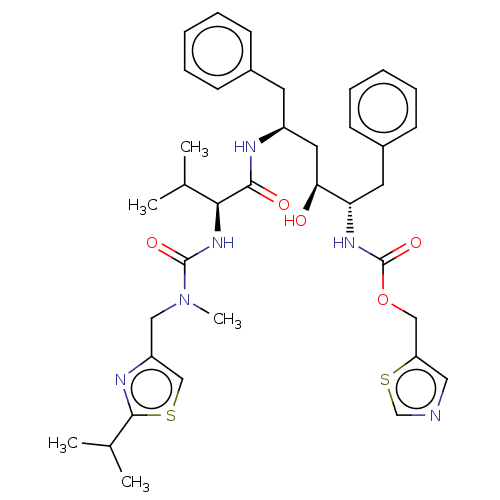
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Liu, H; Xu, L; Hui, H; Vivian, R; Callebaut, C; Murray, BP; Hong, A; Lee, MS; Tsai, LK; Chau, JK; Stray, KM; Cannizzaro, C; Choi, YC; Rhodes, GR; Desai, MC Structure-activity relationships of diamine inhibitors of cytochrome P450 (CYP) 3A as novel pharmacoenhancers, part I: core region. Bioorg Med Chem Lett24:989-94 (2014) [PubMed] Article
Liu, H; Xu, L; Hui, H; Vivian, R; Callebaut, C; Murray, BP; Hong, A; Lee, MS; Tsai, LK; Chau, JK; Stray, KM; Cannizzaro, C; Choi, YC; Rhodes, GR; Desai, MC Structure-activity relationships of diamine inhibitors of cytochrome P450 (CYP) 3A as novel pharmacoenhancers, part I: core region. Bioorg Med Chem Lett24:989-94 (2014) [PubMed] Article