

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Neuronal acetylcholine receptor subunit alpha-7 | ||
| Ligand | BDBM50006621 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1335296 (CHEMBL3238720) | ||
| Ki | 0.930000±n/a nM | ||
| Citation |  Gao, Y; Kellar, KJ; Yasuda, RP; Tran, T; Xiao, Y; Dannals, RF; Horti, AG Derivatives of dibenzothiophene for positron emission tomography imaging ofa7-nicotinic acetylcholine receptors. J Med Chem56:7574-89 (2013) [PubMed] Article Gao, Y; Kellar, KJ; Yasuda, RP; Tran, T; Xiao, Y; Dannals, RF; Horti, AG Derivatives of dibenzothiophene for positron emission tomography imaging ofa7-nicotinic acetylcholine receptors. J Med Chem56:7574-89 (2013) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Neuronal acetylcholine receptor subunit alpha-7 | |||
| Name: | Neuronal acetylcholine receptor subunit alpha-7 | ||
| Synonyms: | ACHA7_RAT | Acra7 | Cholinergic, Nicotinic Alpha7 | Cholinergic, Nicotinic Alpha7/5-HT3 | Chrna7 | Neuronal acetylcholine receptor | Neuronal acetylcholine receptor (alpha7 nAChR) | Neuronal acetylcholine receptor subunit alpha 7 | Neuronal acetylcholine receptor subunit alpha-7 | Neuronal acetylcholine receptor subunit alpha-7 (nAChR alpha7) | Neuronal acetylcholine receptor subunit alpha-7 (nAChR) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 56502.44 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | Q05941 | ||
| Residue: | 502 | ||
| Sequence: |
| ||
| BDBM50006621 | |||
| n/a | |||
| Name | BDBM50006621 | ||
| Synonyms: | CHEMBL3235487 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C19H19IN2O2S | ||
| Mol. Mass. | 466.336 | ||
| SMILES | Ic1cccc2-c3ccc(cc3S(=O)(=O)c12)N1CCN2CCC1CC2 |(29.41,-16.94,;30.31,-18.18,;29.68,-19.6,;30.58,-20.84,;32.11,-20.67,;32.75,-19.26,;34.21,-18.78,;35.54,-19.56,;36.88,-18.79,;36.88,-17.24,;35.54,-16.47,;34.21,-17.24,;32.75,-16.77,;33.07,-15.26,;31.6,-15.74,;31.84,-18.02,;38.22,-16.47,;39.42,-17.43,;40.92,-17.09,;41.58,-15.71,;40.92,-14.31,;39.42,-13.97,;38.22,-14.92,;39.51,-14.95,;40.15,-16.52,)| | ||
| Structure | 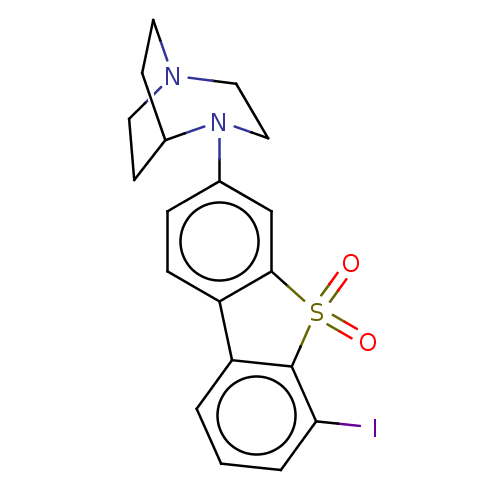
| ||