| Citation |  Ellsworth, BA; Sher, PM; Wu, X; Wu, G; Sulsky, RB; Gu, Z; Murugesan, N; Zhu, Y; Yu, G; Sitkoff, DF; Carlson, KE; Kang, L; Yang, Y; Lee, N; Baska, RA; Keim, WJ; Cullen, MJ; Azzara, AV; Zuvich, E; Thomas, MA; Rohrbach, KW; Devenny, JJ; Godonis, HE; Harvey, SJ; Murphy, BJ; Everlof, GG; Stetsko, PI; Gudmundsson, O; Johnghar, S; Ranasinghe, A; Behnia, K; Pelleymounter, MA; Ewing, WR Reductions in log P improved protein binding and clearance predictions enabling the prospective design of cannabinoid receptor (CB1) antagonists with desired pharmacokinetic properties. J Med Chem56:9586-600 (2014) [PubMed] Article Ellsworth, BA; Sher, PM; Wu, X; Wu, G; Sulsky, RB; Gu, Z; Murugesan, N; Zhu, Y; Yu, G; Sitkoff, DF; Carlson, KE; Kang, L; Yang, Y; Lee, N; Baska, RA; Keim, WJ; Cullen, MJ; Azzara, AV; Zuvich, E; Thomas, MA; Rohrbach, KW; Devenny, JJ; Godonis, HE; Harvey, SJ; Murphy, BJ; Everlof, GG; Stetsko, PI; Gudmundsson, O; Johnghar, S; Ranasinghe, A; Behnia, K; Pelleymounter, MA; Ewing, WR Reductions in log P improved protein binding and clearance predictions enabling the prospective design of cannabinoid receptor (CB1) antagonists with desired pharmacokinetic properties. J Med Chem56:9586-600 (2014) [PubMed] Article |
|---|
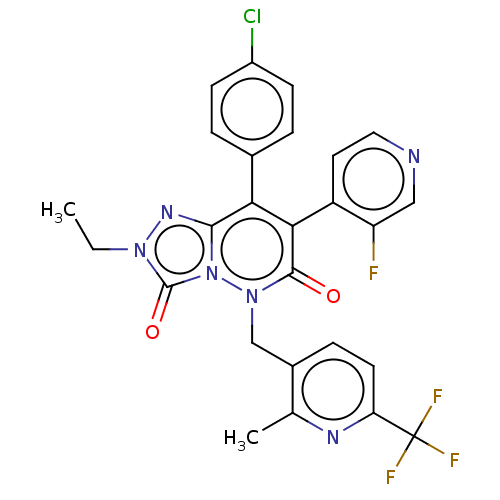
| SMILES | CCn1nc2c(-c3ccc(Cl)cc3)c(-c3ccncc3F)c(=O)n(Cc3ccc(nc3C)C(F)(F)F)n2c1=O |(14.54,-16.67,;15.31,-15.33,;14.53,-14,;13,-13.84,;12.68,-12.33,;11.35,-11.56,;10.01,-12.34,;10.02,-13.88,;8.68,-14.65,;7.35,-13.89,;6.01,-14.66,;7.35,-12.34,;8.68,-11.57,;11.34,-10.03,;10.01,-9.26,;8.68,-10.03,;7.35,-9.26,;7.34,-7.72,;8.67,-6.95,;10.01,-7.72,;11.34,-6.95,;12.68,-9.26,;12.67,-7.72,;14.01,-10.02,;15.35,-9.25,;15.34,-7.71,;14,-6.95,;13.98,-5.41,;15.32,-4.62,;16.67,-5.39,;16.67,-6.93,;18.01,-7.69,;15.31,-3.08,;16.64,-2.3,;13.97,-2.32,;15.3,-1.53,;14.01,-11.57,;15.16,-12.6,;16.66,-12.27,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ellsworth, BA; Sher, PM; Wu, X; Wu, G; Sulsky, RB; Gu, Z; Murugesan, N; Zhu, Y; Yu, G; Sitkoff, DF; Carlson, KE; Kang, L; Yang, Y; Lee, N; Baska, RA; Keim, WJ; Cullen, MJ; Azzara, AV; Zuvich, E; Thomas, MA; Rohrbach, KW; Devenny, JJ; Godonis, HE; Harvey, SJ; Murphy, BJ; Everlof, GG; Stetsko, PI; Gudmundsson, O; Johnghar, S; Ranasinghe, A; Behnia, K; Pelleymounter, MA; Ewing, WR Reductions in log P improved protein binding and clearance predictions enabling the prospective design of cannabinoid receptor (CB1) antagonists with desired pharmacokinetic properties. J Med Chem56:9586-600 (2014) [PubMed] Article
Ellsworth, BA; Sher, PM; Wu, X; Wu, G; Sulsky, RB; Gu, Z; Murugesan, N; Zhu, Y; Yu, G; Sitkoff, DF; Carlson, KE; Kang, L; Yang, Y; Lee, N; Baska, RA; Keim, WJ; Cullen, MJ; Azzara, AV; Zuvich, E; Thomas, MA; Rohrbach, KW; Devenny, JJ; Godonis, HE; Harvey, SJ; Murphy, BJ; Everlof, GG; Stetsko, PI; Gudmundsson, O; Johnghar, S; Ranasinghe, A; Behnia, K; Pelleymounter, MA; Ewing, WR Reductions in log P improved protein binding and clearance predictions enabling the prospective design of cannabinoid receptor (CB1) antagonists with desired pharmacokinetic properties. J Med Chem56:9586-600 (2014) [PubMed] Article