| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Delta-aminolevulinic acid dehydratase, chloroplastic |
|---|
| Ligand | BDBM50017195 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1365592 (CHEMBL3297373) |
|---|
| IC50 | 88200±n/a nM |
|---|
| Citation |  Lentz, CS; Halls, VS; Hannam, JS; Strassel, S; Lawrence, SH; Jaffe, EK; Famulok, M; Hoerauf, A; Pfarr, KM wALADin benzimidazoles differentially modulate the function of porphobilinogen synthase orthologs. J Med Chem57:2498-510 (2014) [PubMed] Article Lentz, CS; Halls, VS; Hannam, JS; Strassel, S; Lawrence, SH; Jaffe, EK; Famulok, M; Hoerauf, A; Pfarr, KM wALADin benzimidazoles differentially modulate the function of porphobilinogen synthase orthologs. J Med Chem57:2498-510 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Delta-aminolevulinic acid dehydratase, chloroplastic |
|---|
| Name: | Delta-aminolevulinic acid dehydratase, chloroplastic |
|---|
| Synonyms: | ALAD | ALADH | HEM2_PEA | HEMB |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 43811.42 |
|---|
| Organism: | Pisum sativum |
|---|
| Description: | ChEMBL_108384 |
|---|
| Residue: | 398 |
|---|
| Sequence: | HTFVDLKSPFTLSNYLSFSSSKRRQPPSLFTVRASDSDFEAAVVAGKVPEAPPVPPTPAS
PAGTPVVPSLPIQRRPRRNRRSPALRSAFQETTLSPANFVYPLFIHEGEEDTPIGAMPGC
YRLGWRHGLLEEVAKARDVGVNSVVLFPKIPDALKTPTGDEAYNEDGLVPRSIRLLKDKY
PDLIIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQARAGADVVSPSDMMD
GRVGAMRVALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFGDKKTYQMNPANYRE
ALTEMREDESEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSGEYSMIKAGGALKMI
DEEKVMMESLLCLRRAGADIILTYFALQAARTLCGEKR
|
|
|
|---|
| BDBM50017195 |
|---|
| n/a |
|---|
| Name | BDBM50017195 |
|---|
| Synonyms: | CHEMBL2430557 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H15F3N2O3 |
|---|
| Mol. Mass. | 400.3506 |
|---|
| SMILES | OC(=O)c1c2CN(CCc2nc2ccccc12)C(=O)c1cccc(c1)C(F)(F)F |
|---|
| Structure | 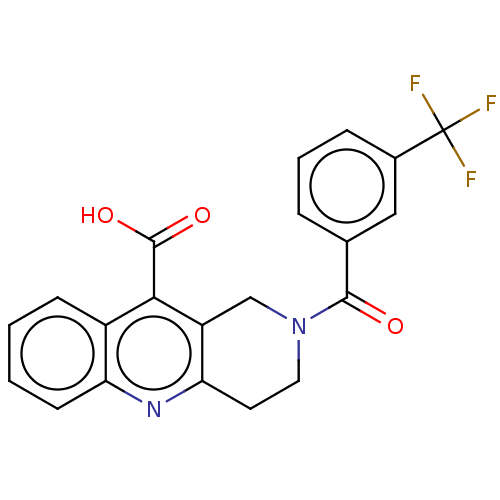
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lentz, CS; Halls, VS; Hannam, JS; Strassel, S; Lawrence, SH; Jaffe, EK; Famulok, M; Hoerauf, A; Pfarr, KM wALADin benzimidazoles differentially modulate the function of porphobilinogen synthase orthologs. J Med Chem57:2498-510 (2014) [PubMed] Article
Lentz, CS; Halls, VS; Hannam, JS; Strassel, S; Lawrence, SH; Jaffe, EK; Famulok, M; Hoerauf, A; Pfarr, KM wALADin benzimidazoles differentially modulate the function of porphobilinogen synthase orthologs. J Med Chem57:2498-510 (2014) [PubMed] Article