| Reaction Details |
|---|
 | Report a problem with these data |
| Target | E3 ubiquitin-protein ligase Mdm2 |
|---|
| Ligand | BDBM50018534 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1362099 (CHEMBL3294652) |
|---|
| Ki | 1370±n/a nM |
|---|
| Citation |  Li, J; Wu, Y; Guo, Z; Zhuang, C; Yao, J; Dong, G; Yu, Z; Min, X; Wang, S; Liu, Y; Wu, S; Zhu, S; Sheng, C; Miao, Z; Zhang, W Discovery of 1-arylpyrrolidone derivatives as potent p53-MDM2 inhibitors based on molecule fusing strategy. Bioorg Med Chem Lett24:2648-50 (2014) [PubMed] Article Li, J; Wu, Y; Guo, Z; Zhuang, C; Yao, J; Dong, G; Yu, Z; Min, X; Wang, S; Liu, Y; Wu, S; Zhu, S; Sheng, C; Miao, Z; Zhang, W Discovery of 1-arylpyrrolidone derivatives as potent p53-MDM2 inhibitors based on molecule fusing strategy. Bioorg Med Chem Lett24:2648-50 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| E3 ubiquitin-protein ligase Mdm2 |
|---|
| Name: | E3 ubiquitin-protein ligase Mdm2 |
|---|
| Synonyms: | Double minute 2 protein | Double minute 2 protein (HDM2) | E3 ubiquitin-protein ligase Mdm2 (p53-binding protein Mdm2) | Hdm2 | Human Double Minute 2 (HDM2) | MDM2 | MDM2-MDMX | MDM2_HUMAN | p53-Binding Protein MDM2 | p53-binding protein |
|---|
| Type: | Oncoprotein |
|---|
| Mol. Mass.: | 55196.54 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q00987 |
|---|
| Residue: | 491 |
|---|
| Sequence: | MCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQY
IMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGT
SVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRRRAISETEENSDELSGERQ
RKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDS
VSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEVYQVTVYQAGESDTDSFEEDPEISLA
DYWKCTSCNEMNPPLPSHCNRCWALRENWLPEDKGKDKGEISEKAKLENSTQAEEGFDVP
DCKKTIVNDSRESCVEENDDKITQASQSQESEDYSQPSTSSSIIYSSQEDVKEFEREETQ
DKEESVESSLPLNAIEPCVICQGRPKNGCIVHGKTGHLMACFTCAKKLKKRNKPCPVCRQ
PIQMIVLTYFP
|
|
|
|---|
| BDBM50018534 |
|---|
| n/a |
|---|
| Name | BDBM50018534 |
|---|
| Synonyms: | CHEMBL3290670 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C32H31BrF3NO4 |
|---|
| Mol. Mass. | 630.492 |
|---|
| SMILES | CC(C)OC1=C(C(N(Cc2ccc(cc2OC(C)(C)C)C(F)(F)F)C1=O)c1ccc(Br)cc1)C(=O)c1ccccc1 |t:4| |
|---|
| Structure | 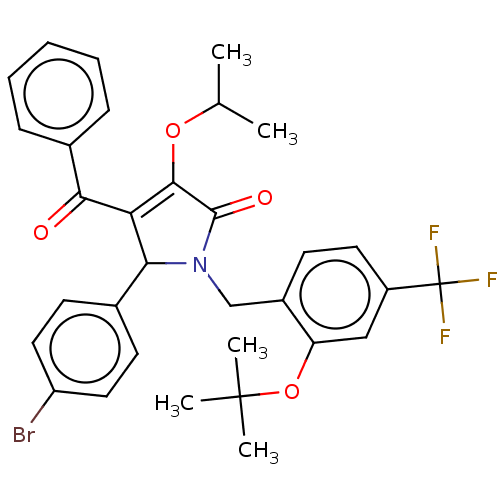
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Li, J; Wu, Y; Guo, Z; Zhuang, C; Yao, J; Dong, G; Yu, Z; Min, X; Wang, S; Liu, Y; Wu, S; Zhu, S; Sheng, C; Miao, Z; Zhang, W Discovery of 1-arylpyrrolidone derivatives as potent p53-MDM2 inhibitors based on molecule fusing strategy. Bioorg Med Chem Lett24:2648-50 (2014) [PubMed] Article
Li, J; Wu, Y; Guo, Z; Zhuang, C; Yao, J; Dong, G; Yu, Z; Min, X; Wang, S; Liu, Y; Wu, S; Zhu, S; Sheng, C; Miao, Z; Zhang, W Discovery of 1-arylpyrrolidone derivatives as potent p53-MDM2 inhibitors based on molecule fusing strategy. Bioorg Med Chem Lett24:2648-50 (2014) [PubMed] Article