

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cytochrome P450 3A4 | ||
| Ligand | BDBM50018769 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1362864 (CHEMBL3291638) | ||
| IC50 | 6100±n/a nM | ||
| Citation |  Gillespie, P; Pietranico-Cole, S; Myers, M; Bilotta, JA; Conde-Knape, K; Fotouhi, N; Goodnow, RA; Guertin, KR; Hamilton, MM; Haynes, NE; Liu, B; Qi, L; Ren, Y; Scott, NR; So, SS; Spence, C; Taub, R; Thakkar, K; Tilley, JW; Zwingelstein, C Discovery of camphor-derived pyrazolones as 11ß-hydroxysteroid dehydrogenase type 1 inhibitors. Bioorg Med Chem Lett24:2707-11 (2014) [PubMed] Article Gillespie, P; Pietranico-Cole, S; Myers, M; Bilotta, JA; Conde-Knape, K; Fotouhi, N; Goodnow, RA; Guertin, KR; Hamilton, MM; Haynes, NE; Liu, B; Qi, L; Ren, Y; Scott, NR; So, SS; Spence, C; Taub, R; Thakkar, K; Tilley, JW; Zwingelstein, C Discovery of camphor-derived pyrazolones as 11ß-hydroxysteroid dehydrogenase type 1 inhibitors. Bioorg Med Chem Lett24:2707-11 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cytochrome P450 3A4 | |||
| Name: | Cytochrome P450 3A4 | ||
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 57349.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 503 | ||
| Sequence: |
| ||
| BDBM50018769 | |||
| n/a | |||
| Name | BDBM50018769 | ||
| Synonyms: | CHEMBL3291346 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C24H24F2N2O | ||
| Mol. Mass. | 394.457 | ||
| SMILES | [H][C@@]12CC[C@@](C)(c3c1c(=O)n(-c1ccc(F)cc1F)n3Cc1ccccc1)C2(C)C |r,wU:4.4,1.0,(19.67,-12.49,;19.67,-10.99,;20.96,-10.25,;20.96,-8.75,;19.67,-8,;19.25,-6.57,;18.38,-8.75,;18.38,-10.25,;16.95,-10.71,;16.5,-12.13,;16.08,-9.5,;14.53,-9.5,;13.77,-10.83,;12.23,-10.84,;11.45,-9.49,;9.91,-9.49,;12.23,-8.16,;13.77,-8.16,;14.55,-6.83,;16.96,-8.29,;16.48,-6.82,;17.51,-5.68,;19.01,-6,;20.04,-4.85,;19.56,-3.38,;18.04,-3.07,;17.02,-4.22,;20.53,-9.49,;22.02,-10.35,;22.02,-8.63,)| | ||
| Structure | 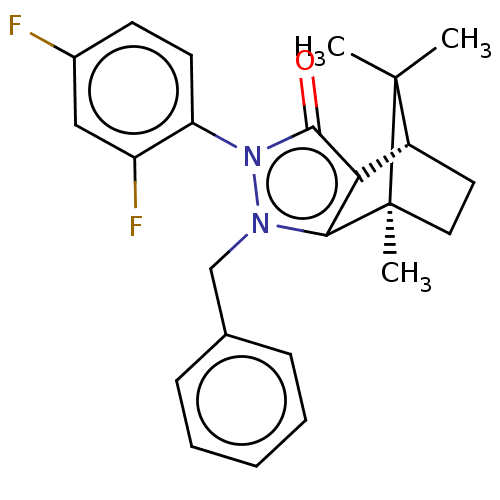
| ||