| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta |
|---|
| Ligand | BDBM18372 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1361547 (CHEMBL3294871) |
|---|
| Kd | 1250±n/a nM |
|---|
| Citation |  Zimmermann, G; Schultz-Fademrecht, C; Küchler, P; Murarka, S; Ismail, S; Triola, G; Nussbaumer, P; Wittinghofer, A; Waldmann, H Structure guided design and kinetic analysis of highly potent benzimidazole inhibitors targeting the PDEd prenyl binding site. J Med Chem57:5435-48 (2014) [PubMed] Article Zimmermann, G; Schultz-Fademrecht, C; Küchler, P; Murarka, S; Ismail, S; Triola, G; Nussbaumer, P; Wittinghofer, A; Waldmann, H Structure guided design and kinetic analysis of highly potent benzimidazole inhibitors targeting the PDEd prenyl binding site. J Med Chem57:5435-48 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta |
|---|
| Name: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | GMP-PDE delta | PDE6D | PDE6D_HUMAN | PDED | Protein p17 | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 17418.30 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_105761 |
|---|
| Residue: | 150 |
|---|
| Sequence: | MSAKDERAREILRGFKLNWMNLRDAETGKILWQGTEDLSVPGVEHEARVPKKILKCKAVS
RELNFSSTEQMEKFRLEQKVYFKGQCLEEWFFEFGFVIPNSTNTWQSLIEAAPESQMMPA
SVLTGNVIIETKFFDDDLLVSTSRVRLFYV
|
|
|
|---|
| BDBM18372 |
|---|
| n/a |
|---|
| Name | BDBM18372 |
|---|
| Synonyms: | (3R,5S,6E)-7-[4-(4-fluorophenyl)-2-(N-methylmethanesulfonamido)-6-(propan-2-yl)pyrimidin-5-yl]-3,5-dihydroxyhept-6-enoic acid | CHEMBL1496 | Ros | Rosuvastatin | US9102656, Rosuvastatin | ZD4522 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H28FN3O6S |
|---|
| Mol. Mass. | 481.538 |
|---|
| SMILES | CC(C)c1nc(nc(-c2ccc(F)cc2)c1\C=C\[C@@H](O)C[C@@H](O)CC(O)=O)N(C)S(C)(=O)=O |r| |
|---|
| Structure | 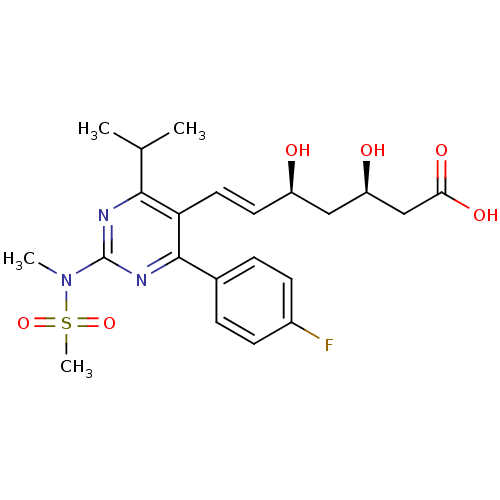
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Zimmermann, G; Schultz-Fademrecht, C; Küchler, P; Murarka, S; Ismail, S; Triola, G; Nussbaumer, P; Wittinghofer, A; Waldmann, H Structure guided design and kinetic analysis of highly potent benzimidazole inhibitors targeting the PDEd prenyl binding site. J Med Chem57:5435-48 (2014) [PubMed] Article
Zimmermann, G; Schultz-Fademrecht, C; Küchler, P; Murarka, S; Ismail, S; Triola, G; Nussbaumer, P; Wittinghofer, A; Waldmann, H Structure guided design and kinetic analysis of highly potent benzimidazole inhibitors targeting the PDEd prenyl binding site. J Med Chem57:5435-48 (2014) [PubMed] Article