

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cytochrome P450 3A4 | ||
| Ligand | BDBM50011134 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1366300 (CHEMBL3297633) | ||
| IC50 | 23000±n/a nM | ||
| Citation |  Fader, LD; Carson, R; Morin, S; Bilodeau, F; Chabot, C; Halmos, T; Bailey, MD; Kawai, SH; Coulombe, R; Laplante, S; Mekhssian, K; Jakalian, A; Garneau, M; Duan, J; Mason, SW; Simoneau, B; Fenwick, C; Tsantrizos, Y; Yoakim, C Minimizing the Contribution of Enterohepatic Recirculation to Clearance in Rat for the NCINI Class of Inhibitors of HIV. ACS Med Chem Lett5:711-6 (2014) [PubMed] Article Fader, LD; Carson, R; Morin, S; Bilodeau, F; Chabot, C; Halmos, T; Bailey, MD; Kawai, SH; Coulombe, R; Laplante, S; Mekhssian, K; Jakalian, A; Garneau, M; Duan, J; Mason, SW; Simoneau, B; Fenwick, C; Tsantrizos, Y; Yoakim, C Minimizing the Contribution of Enterohepatic Recirculation to Clearance in Rat for the NCINI Class of Inhibitors of HIV. ACS Med Chem Lett5:711-6 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cytochrome P450 3A4 | |||
| Name: | Cytochrome P450 3A4 | ||
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 57349.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 503 | ||
| Sequence: |
| ||
| BDBM50011134 | |||
| n/a | |||
| Name | BDBM50011134 | ||
| Synonyms: | CHEMBL3259907 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C27H26N2O4 | ||
| Mol. Mass. | 442.5063 | ||
| SMILES | Cc1nc2ccccc2c(-c2ccc3OCCc4ccnc2c34)c1[C@H](OC(C)(C)C)C(O)=O |r,wD:24.29,(15.05,-15.73,;13.72,-14.97,;12.38,-15.74,;11.05,-14.98,;9.71,-15.75,;8.38,-14.98,;8.38,-13.44,;9.71,-12.67,;11.05,-13.43,;12.37,-12.66,;12.36,-11.13,;13.7,-10.35,;13.69,-8.81,;12.35,-8.06,;12.34,-6.52,;11,-5.76,;9.67,-6.54,;9.68,-8.09,;8.37,-8.86,;8.39,-10.39,;9.72,-11.14,;11.03,-10.37,;11.02,-8.84,;13.71,-13.42,;15.04,-12.64,;15.03,-11.1,;16.36,-10.32,;16.35,-8.78,;17.69,-11.09,;17.69,-9.54,;16.38,-13.4,;16.39,-14.94,;17.71,-12.63,)| | ||
| Structure | 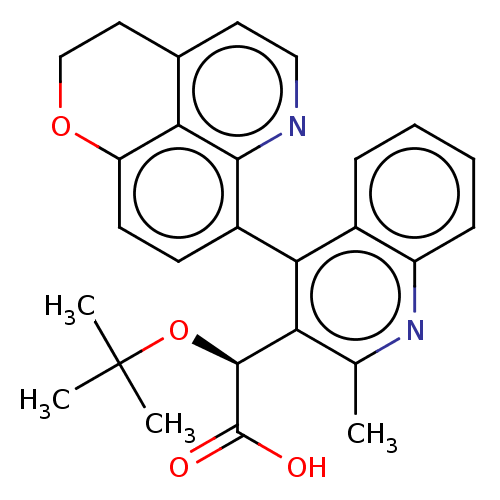
| ||