

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Carbonic anhydrase 9 | ||
| Ligand | BDBM50031534 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1440917 (CHEMBL3375181) | ||
| Ki | 8.5±n/a nM | ||
| Citation |  Bozdag, M; Pinard, M; Carta, F; Masini, E; Scozzafava, A; McKenna, R; Supuran, CT A class of 4-sulfamoylphenyl-¿-aminoalkyl ethers with effective carbonic anhydrase inhibitory action and antiglaucoma effects. J Med Chem57:9673-86 (2014) [PubMed] Article Bozdag, M; Pinard, M; Carta, F; Masini, E; Scozzafava, A; McKenna, R; Supuran, CT A class of 4-sulfamoylphenyl-¿-aminoalkyl ethers with effective carbonic anhydrase inhibitory action and antiglaucoma effects. J Med Chem57:9673-86 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Carbonic anhydrase 9 | |||
| Name: | Carbonic anhydrase 9 | ||
| Synonyms: | CA-IX | CA9 | CAH9_HUMAN | Carbonate dehydratase IX | Carbonic anhydrase 9 (CA IX) | Carbonic anhydrase 9 (CAIX) | Carbonic anhydrase 9 precursor | Carbonic anhydrase IX (CA IX) | Carbonic anhydrase IX (CAIX) | Carbonic anhydrases IX | Carbonic anhydrases; II & IX | G250 | MN | Membrane antigen MN | RCC-associated antigen G250 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 49669.03 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Catalytic domain of human cloned isozyme was used in the assay | ||
| Residue: | 459 | ||
| Sequence: |
| ||
| BDBM50031534 | |||
| n/a | |||
| Name | BDBM50031534 | ||
| Synonyms: | CHEMBL3359174 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C29H23N3O8S2 | ||
| Mol. Mass. | 605.638 | ||
| SMILES | NS(=O)(=O)c1ccc(OCCNC(=S)Nc2ccc(c(c2)C(O)=O)-c2c3ccc(O)cc3oc3cc(=O)ccc23)cc1 |(8.83,2.51,;7.49,1.74,;5.95,1.75,;6.73,3.08,;7.49,.21,;6.15,-.57,;6.15,-2.11,;7.48,-2.89,;7.48,-4.43,;8.82,-5.2,;8.82,-6.74,;10.15,-7.51,;10.22,-9.05,;11.58,-9.77,;8.91,-9.88,;8.98,-11.42,;7.67,-12.25,;7.73,-13.78,;9.09,-14.5,;10.4,-13.67,;10.34,-12.14,;11.76,-14.39,;13.07,-13.57,;10.42,-15.15,;9.15,-16.03,;7.85,-16.85,;6.49,-16.14,;5.2,-16.96,;5.27,-18.49,;3.97,-19.31,;6.62,-19.21,;7.92,-18.39,;9.28,-19.1,;10.57,-18.28,;11.93,-18.99,;13.23,-18.16,;14.58,-18.88,;13.16,-16.63,;11.82,-15.93,;10.5,-16.74,;8.82,-2.11,;8.82,-.57,)| | ||
| Structure | 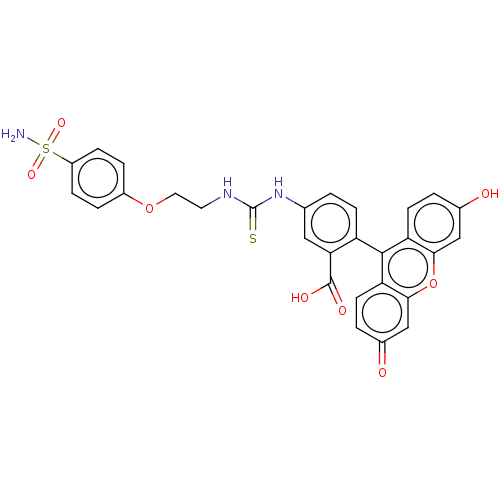
| ||