| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Polyamine deacetylase HDAC10 |
|---|
| Ligand | BDBM50032270 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1440129 (CHEMBL3382542) |
|---|
| Ki | 170±n/a nM |
|---|
| Citation |  Maolanon, AR; Villadsen, JS; Christensen, NJ; Hoeck, C; Friis, T; Harris, P; Gotfredsen, CH; Fristrup, P; Olsen, CA Methyl effect in azumamides provides insight into histone deacetylase inhibition by macrocycles. J Med Chem57:9644-57 (2014) [PubMed] Article Maolanon, AR; Villadsen, JS; Christensen, NJ; Hoeck, C; Friis, T; Harris, P; Gotfredsen, CH; Fristrup, P; Olsen, CA Methyl effect in azumamides provides insight into histone deacetylase inhibition by macrocycles. J Med Chem57:9644-57 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Polyamine deacetylase HDAC10 |
|---|
| Name: | Polyamine deacetylase HDAC10 |
|---|
| Synonyms: | HD10 | HDA10_HUMAN | HDAC10 | Histone deacetylase | Histone deacetylase 10 | Human HDAC10 |
|---|
| Type: | Chromatin regulator; hydrolase; repressor |
|---|
| Mol. Mass.: | 71431.89 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q969S8 |
|---|
| Residue: | 669 |
|---|
| Sequence: | MGTALVYHEDMTATRLLWDDPECEIERPERLTAALDRLRQRGLEQRCLRLSAREASEEEL
GLVHSPEYVSLVRETQVLGKEELQALSGQFDAIYFHPSTFHCARLAAGAGLQLVDAVLTG
AVQNGLALVRPPGHHGQRAAANGFCVFNNVAIAAAHAKQKHGLHRILVVDWDVHHGQGIQ
YLFEDDPSVLYFSWHRYEHGRFWPFLRESDADAVGRGQGLGFTVNLPWNQVGMGNADYVA
AFLHLLLPLAFEFDPELVLVSAGFDSAIGDPEGQMQATPECFAHLTQLLQVLAGGRVCAV
LEGGYHLESLAESVCMTVQTLLGDPAPPLSGPMAPCQSALESIQSARAAQAPHWKSLQQQ
DVTAVPMSPSSHSPEGRPPPLLPGGPVCKAAASAPSSLLDQPCLCPAPSVRTAVALTTPD
ITLVLPPDVIQQEASALREETEAWARPHESLAREEALTALGKLLYLLDGMLDGQVNSGIA
ATPASAAAATLDVAVRRGLSHGAQRLLCVALGQLDRPPDLAHDGRSLWLNIRGKEAAALS
MFHVSTPLPVMTGGFLSCILGLVLPLAYGFQPDLVLVALGPGHGLQGPHAALLAAMLRGL
AGGRVLALLEENSTPQLAGILARVLNGEAPPSLGPSSVASPEDVQALMYLRGQLEPQWKM
LQCHPHLVA
|
|
|
|---|
| BDBM50032270 |
|---|
| n/a |
|---|
| Name | BDBM50032270 |
|---|
| Synonyms: | CHEMBL3352995 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H39N5O6 |
|---|
| Mol. Mass. | 541.6392 |
|---|
| SMILES | CC(C)[C@H]1NC(=O)C[C@@H](CCCCCC(O)=O)NC(=O)[C@@H](Cc2c[nH]c3ccccc23)NC(=O)[C@@H](C)NC1=O |r| |
|---|
| Structure | 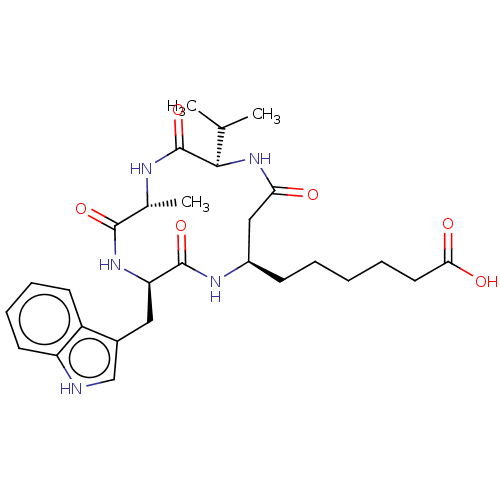
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Maolanon, AR; Villadsen, JS; Christensen, NJ; Hoeck, C; Friis, T; Harris, P; Gotfredsen, CH; Fristrup, P; Olsen, CA Methyl effect in azumamides provides insight into histone deacetylase inhibition by macrocycles. J Med Chem57:9644-57 (2014) [PubMed] Article
Maolanon, AR; Villadsen, JS; Christensen, NJ; Hoeck, C; Friis, T; Harris, P; Gotfredsen, CH; Fristrup, P; Olsen, CA Methyl effect in azumamides provides insight into histone deacetylase inhibition by macrocycles. J Med Chem57:9644-57 (2014) [PubMed] Article