| Reaction Details |
|---|
 | Report a problem with these data |
| Target | UDP-N-acetylmuramoylalanine--D-glutamate ligase |
|---|
| Ligand | BDBM50046818 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1452802 (CHEMBL3365144) |
|---|
| IC50 | 60000±n/a nM |
|---|
| Citation |  Perdih, A; Hrast, M; Barreteau, H; Gobec, S; Wolber, G; Solmajer, T Benzene-1,3-dicarboxylic acid 2,5-dimethylpyrrole derivatives as multiple inhibitors of bacterial Mur ligases (MurC-MurF). Bioorg Med Chem22:4124-34 (2014) [PubMed] Article Perdih, A; Hrast, M; Barreteau, H; Gobec, S; Wolber, G; Solmajer, T Benzene-1,3-dicarboxylic acid 2,5-dimethylpyrrole derivatives as multiple inhibitors of bacterial Mur ligases (MurC-MurF). Bioorg Med Chem22:4124-34 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| UDP-N-acetylmuramoylalanine--D-glutamate ligase |
|---|
| Name: | UDP-N-acetylmuramoylalanine--D-glutamate ligase |
|---|
| Synonyms: | D-glutamic acid-adding enzyme | MURD_ECOLI | MurD (E. coli) | UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase | murD |
|---|
| Type: | Ligase |
|---|
| Mol. Mass.: | 46963.02 |
|---|
| Organism: | Escherichia coli (strain K12) |
|---|
| Description: | n/a |
|---|
| Residue: | 438 |
|---|
| Sequence: | MADYQGKNVVIIGLGLTGLSCVDFFLARGVTPRVMDTRMTPPGLDKLPEAVERHTGSLND
EWLMAADLIVASPGIALAHPSLSAAADAGIEIVGDIELFCREAQAPIVAITGSNGKSTVT
TLVGEMAKAAGVNVGVGGNIGLPALMLLDDECELYVLELSSFQLETTSSLQAVAATILNV
TEDHMDRYPFGLQQYRAAKLRIYENAKVCVVNADDALTMPIRGADERCVSFGVNMGDYHL
NHQQGETWLRVKGEKVLNVKEMKLSGQHNYTNALAALALADAAGLPRASSLKALTTFTGL
PHRFEVVLEHNGVRWINDSKATNVGSTEAALNGLHVDGTLHLLLGGDGKSADFSPLARYL
NGDNVRLYCFGRDGAQLAALRPEVAEQTETMEQAMRLLAPRVQPGDMVLLSPACASLDQF
KNFEQRGNEFARLAKELG
|
|
|
|---|
| BDBM50046818 |
|---|
| n/a |
|---|
| Name | BDBM50046818 |
|---|
| Synonyms: | CHEMBL3310464 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H18ClN3O5S |
|---|
| Mol. Mass. | 495.935 |
|---|
| SMILES | Cc1cc(\C=C2\S\C(NC2=O)=N\c2ccc(Cl)cc2)c(C)n1-c1cc(cc(c1)C(O)=O)C(O)=O |
|---|
| Structure | 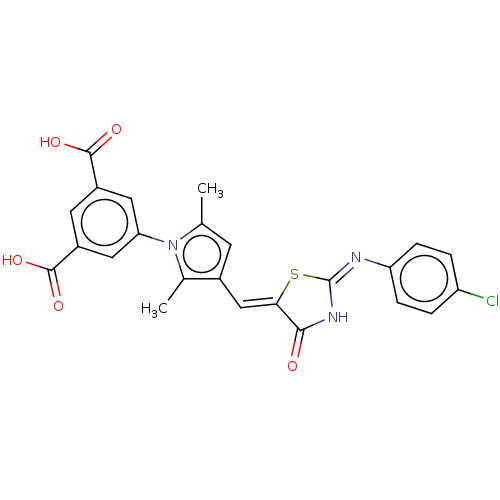
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Perdih, A; Hrast, M; Barreteau, H; Gobec, S; Wolber, G; Solmajer, T Benzene-1,3-dicarboxylic acid 2,5-dimethylpyrrole derivatives as multiple inhibitors of bacterial Mur ligases (MurC-MurF). Bioorg Med Chem22:4124-34 (2014) [PubMed] Article
Perdih, A; Hrast, M; Barreteau, H; Gobec, S; Wolber, G; Solmajer, T Benzene-1,3-dicarboxylic acid 2,5-dimethylpyrrole derivatives as multiple inhibitors of bacterial Mur ligases (MurC-MurF). Bioorg Med Chem22:4124-34 (2014) [PubMed] Article