| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor gamma |
|---|
| Ligand | BDBM24560 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1460054 (CHEMBL3368588) |
|---|
| EC50 | 3600±n/a nM |
|---|
| Citation |  Lamers, C; Dittrich, M; Steri, R; Proschak, E; Schubert-Zsilavecz, M Molecular determinants for improved activity at PPARa: structure-activity relationship of pirinixic acid derivatives, docking study and site-directed mutagenesis of PPARa. Bioorg Med Chem Lett24:4048-52 (2014) [PubMed] Article Lamers, C; Dittrich, M; Steri, R; Proschak, E; Schubert-Zsilavecz, M Molecular determinants for improved activity at PPARa: structure-activity relationship of pirinixic acid derivatives, docking study and site-directed mutagenesis of PPARa. Bioorg Med Chem Lett24:4048-52 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor gamma |
|---|
| Name: | Peroxisome proliferator-activated receptor gamma |
|---|
| Synonyms: | NR1C3 | Nuclear receptor subfamily 1 group C member 3 | PPAR-gamma | PPARG | PPARG_HUMAN | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor gamma (PPAR gamma) | Peroxisome proliferator-activated receptor gamma (PPARG) | Peroxisome proliferator-activated receptor gamma (PPARγ) | Peroxisome proliferator-activated receptor gamma/Nuclear receptor corepressor 2 | peroxisome proliferator-activated receptor gamma isoform 2 |
|---|
| Type: | Nuclear Receptor |
|---|
| Mol. Mass.: | 57613.46 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P37231 |
|---|
| Residue: | 505 |
|---|
| Sequence: | MGETLGDSPIDPESDSFTDTLSANISQEMTMVDTEMPFWPTNFGISSVDLSVMEDHSHSF
DIKPFTTVDFSSISTPHYEDIPFTRTDPVVADYKYDLKLQEYQSAIKVEPASPPYYSEKT
QLYNKPHEEPSNSLMAIECRVCGDKASGFHYGVHACEGCKGFFRRTIRLKLIYDRCDLNC
RIHKKSRNKCQYCRFQKCLAVGMSHNAIRFGRMPQAEKEKLLAEISSDIDQLNPESADLR
ALAKHLYDSYIKSFPLTKAKARAILTGKTTDKSPFVIYDMNSLMMGEDKIKFKHITPLQE
QSKEVAIRIFQGCQFRSVEAVQEITEYAKSIPGFVNLDLNDQVTLLKYGVHEIIYTMLAS
LMNKDGVLISEGQGFMTREFLKSLRKPFGDFMEPKFEFAVKFNALELDDSDLAIFIAVII
LSGDRPGLLNVKPIEDIQDNLLQALELQLKLNHPESSQLFAKLLQKMTDLRQIVTEHVQL
LQVIKKTETDMSLHPLLQEIYKDLY
|
|
|
|---|
| BDBM24560 |
|---|
| n/a |
|---|
| Name | BDBM24560 |
|---|
| Synonyms: | 2-({4-chloro-6-[(2,3-dimethylphenyl)amino]pyrimidin-2-yl}sulfanyl)octanoic acid | CHEMBL518038 | Pirinixic acid-based compound, 6a |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H26ClN3O2S |
|---|
| Mol. Mass. | 407.957 |
|---|
| SMILES | CCCCCCC(Sc1nc(Cl)cc(Nc2cccc(C)c2C)n1)C(O)=O |
|---|
| Structure | 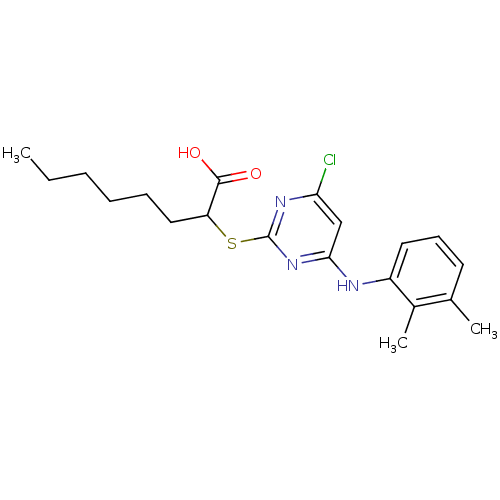
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lamers, C; Dittrich, M; Steri, R; Proschak, E; Schubert-Zsilavecz, M Molecular determinants for improved activity at PPARa: structure-activity relationship of pirinixic acid derivatives, docking study and site-directed mutagenesis of PPARa. Bioorg Med Chem Lett24:4048-52 (2014) [PubMed] Article
Lamers, C; Dittrich, M; Steri, R; Proschak, E; Schubert-Zsilavecz, M Molecular determinants for improved activity at PPARa: structure-activity relationship of pirinixic acid derivatives, docking study and site-directed mutagenesis of PPARa. Bioorg Med Chem Lett24:4048-52 (2014) [PubMed] Article