| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 4 |
|---|
| Ligand | BDBM50051031 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1460073 (CHEMBL3368833) |
|---|
| IC50 | 3.0±n/a nM |
|---|
| Citation |  Islam, MN; Islam, MS; Hoque, MA; Kato, T; Nishino, N; Ito, A; Yoshida, M Bicyclic tetrapeptides as potent HDAC inhibitors: effect of aliphatic loop position and hydrophobicity on inhibitory activity. Bioorg Med Chem22:3862-70 (2014) [PubMed] Article Islam, MN; Islam, MS; Hoque, MA; Kato, T; Nishino, N; Ito, A; Yoshida, M Bicyclic tetrapeptides as potent HDAC inhibitors: effect of aliphatic loop position and hydrophobicity on inhibitory activity. Bioorg Med Chem22:3862-70 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 4 |
|---|
| Name: | Histone deacetylase 4 |
|---|
| Synonyms: | Cereblon/Histone deacetylase 4 | HD4 | HDAC4 | HDAC4_HUMAN | Histone acetylase 4(HDAC4) | Human HDAC4 | KIAA0288 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 119049.39 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P56524 |
|---|
| Residue: | 1084 |
|---|
| Sequence: | MSSQSHPDGLSGRDQPVELLNPARVNHMPSTVDVATALPLQVAPSAVPMDLRLDHQFSLP
VAEPALREQQLQQELLALKQKQQIQRQILIAEFQRQHEQLSRQHEAQLHEHIKQQQEMLA
MKHQQELLEHQRKLERHRQEQELEKQHREQKLQQLKNKEKGKESAVASTEVKMKLQEFVL
NKKKALAHRNLNHCISSDPRYWYGKTQHSSLDQSSPPQSGVSTSYNHPVLGMYDAKDDFP
LRKTASEPNLKLRSRLKQKVAERRSSPLLRRKDGPVVTALKKRPLDVTDSACSSAPGSGP
SSPNNSSGSVSAENGIAPAVPSIPAETSLAHRLVAREGSAAPLPLYTSPSLPNITLGLPA
TGPSAGTAGQQDAERLTLPALQQRLSLFPGTHLTPYLSTSPLERDGGAAHSPLLQHMVLL
EQPPAQAPLVTGLGALPLHAQSLVGADRVSPSIHKLRQHRPLGRTQSAPLPQNAQALQHL
VIQQQHQQFLEKHKQQFQQQQLQMNKIIPKPSEPARQPESHPEETEEELREHQALLDEPY
LDRLPGQKEAHAQAGVQVKQEPIESDEEEAEPPREVEPGQRQPSEQELLFRQQALLLEQQ
RIHQLRNYQASMEAAGIPVSFGGHRPLSRAQSSPASATFPVSVQEPPTKPRFTTGLVYDT
LMLKHQCTCGSSSSHPEHAGRIQSIWSRLQETGLRGKCECIRGRKATLEELQTVHSEAHT
LLYGTNPLNRQKLDSKKLLGSLASVFVRLPCGGVGVDSDTIWNEVHSAGAARLAVGCVVE
LVFKVATGELKNGFAVVRPPGHHAEESTPMGFCYFNSVAVAAKLLQQRLSVSKILIVDWD
VHHGNGTQQAFYSDPSVLYMSLHRYDDGNFFPGSGAPDEVGTGPGVGFNVNMAFTGGLDP
PMGDAEYLAAFRTVVMPIASEFAPDVVLVSSGFDAVEGHPTPLGGYNLSARCFGYLTKQL
MGLAGGRIVLALEGGHDLTAICDASEACVSALLGNELDPLPEKVLQQRPNANAVRSMEKV
MEIHSKYWRCLQRTTSTAGRSLIEAQTCENEEAETVTAMASLSVGVKPAEKRPDEEPMEE
EPPL
|
|
|
|---|
| BDBM50051031 |
|---|
| n/a |
|---|
| Name | BDBM50051031 |
|---|
| Synonyms: | CHEMBL2158745 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C29H43N5O7 |
|---|
| Mol. Mass. | 573.681 |
|---|
| SMILES | [H][C@]12CCCN1C(=O)[C@@]([H])(NC(=O)[C@@H](Cc1ccc(OC)cc1)NC(=O)[C@H](CCCCCC(=O)NO)NC2=O)[C@@H](C)CC |r| |
|---|
| Structure | 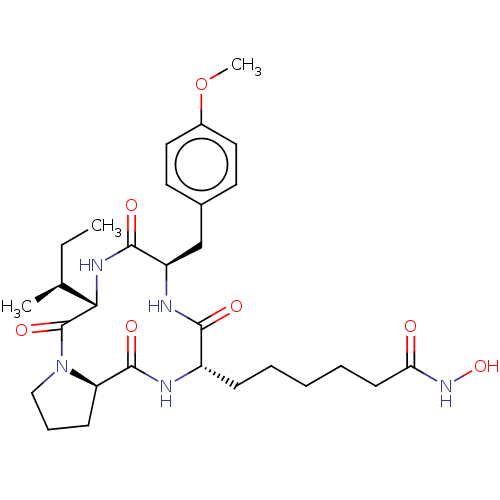
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Islam, MN; Islam, MS; Hoque, MA; Kato, T; Nishino, N; Ito, A; Yoshida, M Bicyclic tetrapeptides as potent HDAC inhibitors: effect of aliphatic loop position and hydrophobicity on inhibitory activity. Bioorg Med Chem22:3862-70 (2014) [PubMed] Article
Islam, MN; Islam, MS; Hoque, MA; Kato, T; Nishino, N; Ito, A; Yoshida, M Bicyclic tetrapeptides as potent HDAC inhibitors: effect of aliphatic loop position and hydrophobicity on inhibitory activity. Bioorg Med Chem22:3862-70 (2014) [PubMed] Article