| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine/threonine-protein kinase PLK3 |
|---|
| Ligand | BDBM50053304 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1459295 (CHEMBL3367525) |
|---|
| IC50 | >50000±n/a nM |
|---|
| Citation |  Laufer, R; Ng, G; Liu, Y; Patel, NK; Edwards, LG; Lang, Y; Li, SW; Feher, M; Awrey, DE; Leung, G; Beletskaya, I; Plotnikova, O; Mason, JM; Hodgson, R; Wei, X; Mao, G; Luo, X; Huang, P; Green, E; Kiarash, R; Lin, DC; Harris-Brandts, M; Ban, F; Nadeem, V; Mak, TW; Pan, GJ; Qiu, W; Chirgadze, NY; Pauls, HW Discovery of inhibitors of the mitotic kinase TTK based on N-(3-(3-sulfamoylphenyl)-1H-indazol-5-yl)-acetamides and carboxamides. Bioorg Med Chem22:4968-97 (2014) [PubMed] Article Laufer, R; Ng, G; Liu, Y; Patel, NK; Edwards, LG; Lang, Y; Li, SW; Feher, M; Awrey, DE; Leung, G; Beletskaya, I; Plotnikova, O; Mason, JM; Hodgson, R; Wei, X; Mao, G; Luo, X; Huang, P; Green, E; Kiarash, R; Lin, DC; Harris-Brandts, M; Ban, F; Nadeem, V; Mak, TW; Pan, GJ; Qiu, W; Chirgadze, NY; Pauls, HW Discovery of inhibitors of the mitotic kinase TTK based on N-(3-(3-sulfamoylphenyl)-1H-indazol-5-yl)-acetamides and carboxamides. Bioorg Med Chem22:4968-97 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Serine/threonine-protein kinase PLK3 |
|---|
| Name: | Serine/threonine-protein kinase PLK3 |
|---|
| Synonyms: | CNK | Cytokine-inducible serine/threonine-protein kinase | FGF-inducible kinase | FNK | PLK3 | PLK3_HUMAN | PRK | Polo-Like Kinase 3 | Proliferation-related kinase | Serine/threonine-protein kinase PLK3 |
|---|
| Type: | Serine/threonine-protein kinase |
|---|
| Mol. Mass.: | 71655.17 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Enzyme assays were using PLK3 purified from baculovirus-infected Trichoplusia ni cells expressing full-length PLK3. |
|---|
| Residue: | 646 |
|---|
| Sequence: | MEPAAGFLSPRPFQRAAAAPAPPAGPGPPPSALRGPELEMLAGLPTSDPGRLITDPRSGR
TYLKGRLLGKGGFARCYEATDTETGSAYAVKVIPQSRVAKPHQREKILNEIELHRDLQHR
HIVRFSHHFEDADNIYIFLELCSRKSLAHIWKARHTLLEPEVRYYLRQILSGLKYLHQRG
ILHRDLKLGNFFITENMELKVGDFGLAARLEPPEQRKKTICGTPNYVAPEVLLRQGHGPE
ADVWSLGCVMYTLLCGSPPFETADLKETYRCIKQVHYTLPASLSLPARQLLAAILRASPR
DRPSIDQILRHDFFTKGYTPDRLPISSCVTVPDLTPPNPARSLFAKVTKSLFGRKKKSKN
HAQERDEVSGLVSGLMRTSVGHQDARPEAPAASGPAPVSLVETAPEDSSPRGTLASSGDG
FEEGLTVATVVESALCALRNCIAFMPPAEQNPAPLAQPEPLVWVSKWVDYSNKFGFGYQL
SSRRVAVLFNDGTHMALSANRKTVHYNPTSTKHFSFSVGAVPRALQPQLGILRYFASYME
QHLMKGGDLPSVEEVEVPAPPLLLQWVKTDQALLMLFSDGTVQVNFYGDHTKLILSGWEP
LLVTFVARNRSACTYLASHLRQLGCSPDLRQRLRYALRLLRDRSPA
|
|
|
|---|
| BDBM50053304 |
|---|
| n/a |
|---|
| Name | BDBM50053304 |
|---|
| Synonyms: | CHEMBL3330410 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H23N5O3S2 |
|---|
| Mol. Mass. | 481.59 |
|---|
| SMILES | NS(=O)(=O)c1cccc(c1)-c1n[nH]c2ccc(NC(=O)C(N3CCCC3)c3ccsc3)cc12 |
|---|
| Structure | 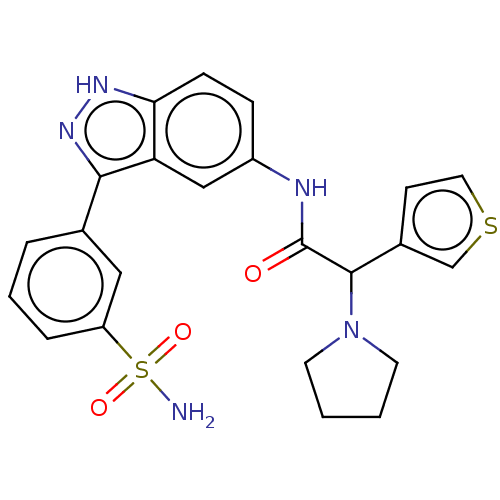
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Laufer, R; Ng, G; Liu, Y; Patel, NK; Edwards, LG; Lang, Y; Li, SW; Feher, M; Awrey, DE; Leung, G; Beletskaya, I; Plotnikova, O; Mason, JM; Hodgson, R; Wei, X; Mao, G; Luo, X; Huang, P; Green, E; Kiarash, R; Lin, DC; Harris-Brandts, M; Ban, F; Nadeem, V; Mak, TW; Pan, GJ; Qiu, W; Chirgadze, NY; Pauls, HW Discovery of inhibitors of the mitotic kinase TTK based on N-(3-(3-sulfamoylphenyl)-1H-indazol-5-yl)-acetamides and carboxamides. Bioorg Med Chem22:4968-97 (2014) [PubMed] Article
Laufer, R; Ng, G; Liu, Y; Patel, NK; Edwards, LG; Lang, Y; Li, SW; Feher, M; Awrey, DE; Leung, G; Beletskaya, I; Plotnikova, O; Mason, JM; Hodgson, R; Wei, X; Mao, G; Luo, X; Huang, P; Green, E; Kiarash, R; Lin, DC; Harris-Brandts, M; Ban, F; Nadeem, V; Mak, TW; Pan, GJ; Qiu, W; Chirgadze, NY; Pauls, HW Discovery of inhibitors of the mitotic kinase TTK based on N-(3-(3-sulfamoylphenyl)-1H-indazol-5-yl)-acetamides and carboxamides. Bioorg Med Chem22:4968-97 (2014) [PubMed] Article