| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
|---|
| Ligand | BDBM18137 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1455848 (CHEMBL3362302) |
|---|
| Ki | 40000±n/a nM |
|---|
| Citation |  Amiable, C; Paoletti, J; Haouz, A; Padilla, A; Labesse, G; Kaminski, PA; Pochet, S 6-(Hetero)Arylpurine nucleotides as inhibitors of the oncogenic target DNPH1: synthesis, structural studies and cytotoxic activities. Eur J Med Chem85:418-37 (2014) [PubMed] Article Amiable, C; Paoletti, J; Haouz, A; Padilla, A; Labesse, G; Kaminski, PA; Pochet, S 6-(Hetero)Arylpurine nucleotides as inhibitors of the oncogenic target DNPH1: synthesis, structural studies and cytotoxic activities. Eur J Med Chem85:418-37 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
|---|
| Name: | 2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
|---|
| Synonyms: | DNPH1_RAT | Deoxyribonucleoside 5'-monophosphate N-glycosidase (Rcl) | Deoxyribonucleoside 5'-monophosphate N-glycosidase (Rcl) D69N | Deoxyribonucleoside 5'-monophosphate N-glycosidase (Rcl) E93Q | Deoxyribonucleoside 5'-monophosphate N-glycosidase (Rcl) S117A | Deoxyribonucleoside 5'-monophosphate N-glycosidase (Rcl) Y13F | Dnph1 | Rcl | c-Myc-responsive protein Rcl |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 17775.23 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | O35820 |
|---|
| Residue: | 163 |
|---|
| Sequence: | MAASGEQAPCSVYFCGSIRGGREDQALYARIVSRLRRYGKVLTEHVADAELEPLGEEAAG
GDQFIHEQDLNWLQQADVVVAEVTQPSLGVGYELGRAVALGKPILCLFRPQSGRVLSAMI
RGAADGSRFQVWDYAEGEVETMLDRYFEAYLPQKTASSSHPSA
|
|
|
|---|
| BDBM18137 |
|---|
| n/a |
|---|
| Name | BDBM18137 |
|---|
| Synonyms: | AMP | CHEMBL752 | US11185100, TABLE 7.3 | [(2R,3S,4R,5R)-5-adenin-9-yl-3,4-dihydroxy-tetrahydrofuran-2-yl]methyl dihydrogen phosphate;hydrate | adenosine 5 -monophosphate | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}phosphonic acid |
|---|
| Type | Nucleoside or nucleotide |
|---|
| Emp. Form. | C10H14N5O7P |
|---|
| Mol. Mass. | 347.2212 |
|---|
| SMILES | Nc1ncnc2n(cnc12)[C@@H]1O[C@H](COP(O)(O)=O)[C@@H](O)[C@H]1O |
|---|
| Structure | 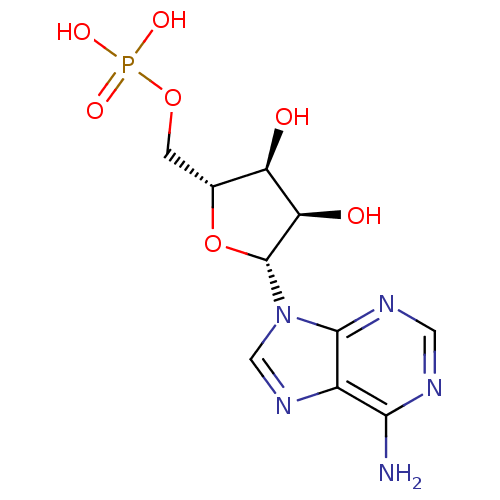
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Amiable, C; Paoletti, J; Haouz, A; Padilla, A; Labesse, G; Kaminski, PA; Pochet, S 6-(Hetero)Arylpurine nucleotides as inhibitors of the oncogenic target DNPH1: synthesis, structural studies and cytotoxic activities. Eur J Med Chem85:418-37 (2014) [PubMed] Article
Amiable, C; Paoletti, J; Haouz, A; Padilla, A; Labesse, G; Kaminski, PA; Pochet, S 6-(Hetero)Arylpurine nucleotides as inhibitors of the oncogenic target DNPH1: synthesis, structural studies and cytotoxic activities. Eur J Med Chem85:418-37 (2014) [PubMed] Article