| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform |
|---|
| Ligand | BDBM50059635 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1460636 (CHEMBL3395877) |
|---|
| IC50 | 25±n/a nM |
|---|
| Citation |  Andrs, M; Korabecny, J; Jun, D; Hodny, Z; Bartek, J; Kuca, K Phosphatidylinositol 3-Kinase (PI3K) and phosphatidylinositol 3-kinase-related kinase (PIKK) inhibitors: importance of the morpholine ring. J Med Chem58:41-71 (2015) [PubMed] Article Andrs, M; Korabecny, J; Jun, D; Hodny, Z; Bartek, J; Kuca, K Phosphatidylinositol 3-Kinase (PI3K) and phosphatidylinositol 3-kinase-related kinase (PIKK) inhibitors: importance of the morpholine ring. J Med Chem58:41-71 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform |
|---|
| Name: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform |
|---|
| Synonyms: | PI3-kinase p110 subunit gamma | PI3-kinase subunit p120-gamma | PI3Kgamma | PIK3CG | PK3CG_HUMAN | Phosphatidylinositol 4,5-biphosphate 3-kinase catalytic subunit gamma (PIK3CG) | Phosphatidylinositol 4,5-bisphosphate 3-kinase (PI3K) | Phosphatidylinositol 4,5-bisphosphate 3-kinase 110 kDa catalytic subunit gamma (PI3K gamma) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma (PI3Kgamma) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (PI3K gamma) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (PI3K) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (PI3Kgamma) | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | Phosphoinositide 3-Kinase (PI3K), gamma Chain A | Phosphoinositide 3-kinases gamma (PI3K gamma) | Phosphoinositide-3-kinase (PI3K gamma) | p120-PI3K |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 126470.30 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P48736 |
|---|
| Residue: | 1102 |
|---|
| Sequence: | MELENYKQPVVLREDNCRRRRRMKPRSAAASLSSMELIPIEFVLPTSQRKCKSPETALLH
VAGHGNVEQMKAQVWLRALETSVAADFYHRLGPHHFLLLYQKKGQWYEIYDKYQVVQTLD
CLRYWKATHRSPGQIHLVQRHPPSEESQAFQRQLTALIGYDVTDVSNVHDDELEFTRRGL
VTPRMAEVASRDPKLYAMHPWVTSKPLPEYLWKKIANNCIFIVIHRSTTSQTIKVSPDDT
PGAILQSFFTKMAKKKSLMDIPESQSEQDFVLRVCGRDEYLVGETPIKNFQWVRHCLKNG
EEIHVVLDTPPDPALDEVRKEEWPLVDDCTGVTGYHEQLTIHGKDHESVFTVSLWDCDRK
FRVKIRGIDIPVLPRNTDLTVFVEANIQHGQQVLCQRRTSPKPFTEEVLWNVWLEFSIKI
KDLPKGALLNLQIYCGKAPALSSKASAESPSSESKGKVQLLYYVNLLLIDHRFLLRRGEY
VLHMWQISGKGEDQGSFNADKLTSATNPDKENSMSISILLDNYCHPIALPKHQPTPDPEG
DRVRAEMPNQLRKQLEAIIATDPLNPLTAEDKELLWHFRYESLKHPKAYPKLFSSVKWGQ
QEIVAKTYQLLARREVWDQSALDVGLTMQLLDCNFSDENVRAIAVQKLESLEDDDVLHYL
LQLVQAVKFEPYHDSALARFLLKRGLRNKRIGHFLFWFLRSEIAQSRHYQQRFAVILEAY
LRGCGTAMLHDFTQQVQVIEMLQKVTLDIKSLSAEKYDVSSQVISQLKQKLENLQNSQLP
ESFRVPYDPGLKAGALAIEKCKVMASKKKPLWLEFKCADPTALSNETIGIIFKHGDDLRQ
DMLILQILRIMESIWETESLDLCLLPYGCISTGDKIGMIEIVKDATTIAKIQQSTVGNTG
AFKDEVLNHWLKEKSPTEEKFQAAVERFVYSCAGYCVATFVLGIGDRHNDNIMITETGNL
FHIDFGHILGNYKSFLGINKERVPFVLTPDFLFVMGTSGKKTSPHFQKFQDICVKAYLAL
RHHTNLLIILFSMMLMTGMPQLTSKEDIEYIRDALTVGKNEEDAKKYFLDQIEVCRDKGW
TVQFNWFLHLVLGIKQGEKHSA
|
|
|
|---|
| BDBM50059635 |
|---|
| n/a |
|---|
| Name | BDBM50059635 |
|---|
| Synonyms: | CHEMBL3393066 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H22N8O |
|---|
| Mol. Mass. | 354.4096 |
|---|
| SMILES | CC(C)n1c(C)nc2c(nc(nc12)N1CCOCC1)-c1cnc(N)nc1 |
|---|
| Structure | 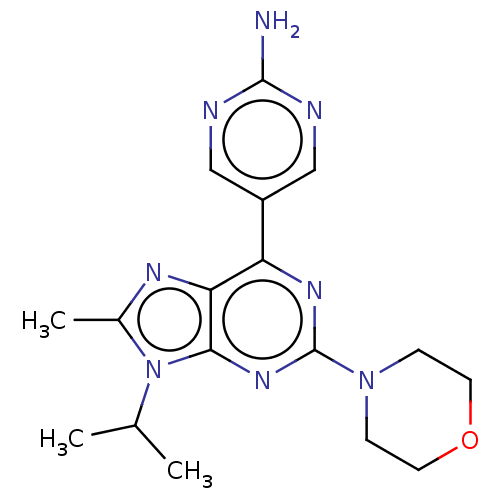
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Andrs, M; Korabecny, J; Jun, D; Hodny, Z; Bartek, J; Kuca, K Phosphatidylinositol 3-Kinase (PI3K) and phosphatidylinositol 3-kinase-related kinase (PIKK) inhibitors: importance of the morpholine ring. J Med Chem58:41-71 (2015) [PubMed] Article
Andrs, M; Korabecny, J; Jun, D; Hodny, Z; Bartek, J; Kuca, K Phosphatidylinositol 3-Kinase (PI3K) and phosphatidylinositol 3-kinase-related kinase (PIKK) inhibitors: importance of the morpholine ring. J Med Chem58:41-71 (2015) [PubMed] Article