| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Beta-secretase 1 |
|---|
| Ligand | BDBM50061593 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1461646 (CHEMBL3395115) |
|---|
| IC50 | 0.900000±n/a nM |
|---|
| Citation |  Chen, JJ; Liu, Q; Yuan, C; Gore, V; Lopez, P; Ma, V; Amegadzie, A; Qian, W; Judd, TC; Minatti, AE; Brown, J; Cheng, Y; Xue, M; Zhong, W; Dineen, TA; Epstein, O; Human, J; Kreiman, C; Marx, I; Weiss, MM; Hitchcock, SA; Powers, TS; Chen, K; Wen, PH; Whittington, DA; Cheng, AC; Bartberger, MD; Hickman, D; Werner, JA; Vargas, HM; Everds, NE; Vonderfecht, SL; Dunn, RT; Wood, S; Fremeau, RT; White, RD; Patel, VF Development of 2-aminooxazoline 3-azaxanthenes as orally efficacious▀-secretase inhibitors for the potential treatment of Alzheimer's disease. Bioorg Med Chem Lett25:767-74 (2015) [PubMed] Article Chen, JJ; Liu, Q; Yuan, C; Gore, V; Lopez, P; Ma, V; Amegadzie, A; Qian, W; Judd, TC; Minatti, AE; Brown, J; Cheng, Y; Xue, M; Zhong, W; Dineen, TA; Epstein, O; Human, J; Kreiman, C; Marx, I; Weiss, MM; Hitchcock, SA; Powers, TS; Chen, K; Wen, PH; Whittington, DA; Cheng, AC; Bartberger, MD; Hickman, D; Werner, JA; Vargas, HM; Everds, NE; Vonderfecht, SL; Dunn, RT; Wood, S; Fremeau, RT; White, RD; Patel, VF Development of 2-aminooxazoline 3-azaxanthenes as orally efficacious▀-secretase inhibitors for the potential treatment of Alzheimer's disease. Bioorg Med Chem Lett25:767-74 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Beta-secretase 1 |
|---|
| Name: | Beta-secretase 1 |
|---|
| Synonyms: | ASP2 | Asp 2 | Aspartyl protease 2 | BACE | BACE1 | BACE1_HUMAN | Beta-secretase (BACE) | Beta-secretase 1 | Beta-secretase 1 (BACE 1) | Beta-secretase 1 (BACE-1) | Beta-secretase 1 (BACE1) | Beta-site APP cleaving enzyme 1 | Beta-site amyloid precursor protein cleaving enzyme 1 | KIAA1149 | Memapsin-2 | Membrane-associated aspartic protease 2 | beta-Secretase (BACE-1) | beta-Secretase (BACE1) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 55755.10 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P56817 |
|---|
| Residue: | 501 |
|---|
| Sequence: | MAQALPWLLLWMGAGVLPAHGTQHGIRLPLRSGLGGAPLGLRLPRETDEEPEEPGRRGSF
VEMVDNLRGKSGQGYYVEMTVGSPPQTLNILVDTGSSNFAVGAAPHPFLHRYYQRQLSST
YRDLRKGVYVPYTQGKWEGELGTDLVSIPHGPNVTVRANIAAITESDKFFINGSNWEGIL
GLAYAEIARPDDSLEPFFDSLVKQTHVPNLFSLQLCGAGFPLNQSEVLASVGGSMIIGGI
DHSLYTGSLWYTPIRREWYYEVIIVRVEINGQDLKMDCKEYNYDKSIVDSGTTNLRLPKK
VFEAAVKSIKAASSTEKFPDGFWLGEQLVCWQAGTTPWNIFPVISLYLMGEVTNQSFRIT
ILPQQYLRPVEDVATSQDDCYKFAISQSSTGTVMGAVIMEGFYVVFDRARKRIGFAVSAC
HVHDEFRTAAVEGPFVTLDMEDCGYNIPQTDESTLMTIAYVMAAICALFMLPLCLMVCQW
RCLRCLRQQHDDFADDISLLK
|
|
|
|---|
| BDBM50061593 |
|---|
| n/a |
|---|
| Name | BDBM50061593 |
|---|
| Synonyms: | CHEMBL3394040 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H23FN4O3 |
|---|
| Mol. Mass. | 458.4842 |
|---|
| SMILES | CC1(C)OCCC(=C1)c1cc2c(Oc3ccc(cc3[C@@]22COC(N)=N2)-c2cccnc2F)cn1 |r,c:6,26| |
|---|
| Structure | 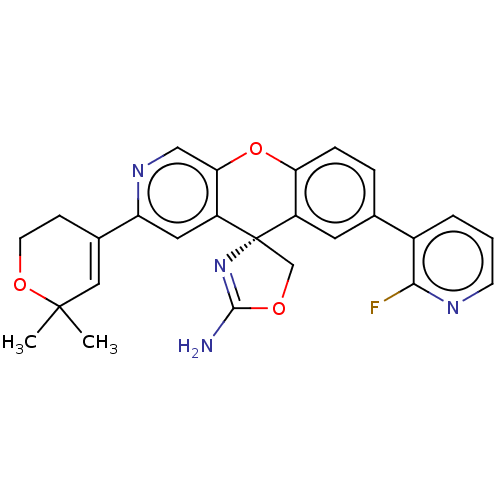
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Chen, JJ; Liu, Q; Yuan, C; Gore, V; Lopez, P; Ma, V; Amegadzie, A; Qian, W; Judd, TC; Minatti, AE; Brown, J; Cheng, Y; Xue, M; Zhong, W; Dineen, TA; Epstein, O; Human, J; Kreiman, C; Marx, I; Weiss, MM; Hitchcock, SA; Powers, TS; Chen, K; Wen, PH; Whittington, DA; Cheng, AC; Bartberger, MD; Hickman, D; Werner, JA; Vargas, HM; Everds, NE; Vonderfecht, SL; Dunn, RT; Wood, S; Fremeau, RT; White, RD; Patel, VF Development of 2-aminooxazoline 3-azaxanthenes as orally efficacious▀-secretase inhibitors for the potential treatment of Alzheimer's disease. Bioorg Med Chem Lett25:767-74 (2015) [PubMed] Article
Chen, JJ; Liu, Q; Yuan, C; Gore, V; Lopez, P; Ma, V; Amegadzie, A; Qian, W; Judd, TC; Minatti, AE; Brown, J; Cheng, Y; Xue, M; Zhong, W; Dineen, TA; Epstein, O; Human, J; Kreiman, C; Marx, I; Weiss, MM; Hitchcock, SA; Powers, TS; Chen, K; Wen, PH; Whittington, DA; Cheng, AC; Bartberger, MD; Hickman, D; Werner, JA; Vargas, HM; Everds, NE; Vonderfecht, SL; Dunn, RT; Wood, S; Fremeau, RT; White, RD; Patel, VF Development of 2-aminooxazoline 3-azaxanthenes as orally efficacious▀-secretase inhibitors for the potential treatment of Alzheimer's disease. Bioorg Med Chem Lett25:767-74 (2015) [PubMed] Article