| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone-lysine N-methyltransferase EZH1 |
|---|
| Ligand | BDBM50009672 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1463671 (CHEMBL3399058) |
|---|
| IC50 | 6400±n/a nM |
|---|
| Citation |  Kung, PP; Huang, B; Zehnder, L; Tatlock, J; Bingham, P; Krivacic, C; Gajiwala, K; Diehl, W; Yu, X; Maegley, KA SAH derived potent and selective EZH2 inhibitors. Bioorg Med Chem Lett25:1532-7 (2015) [PubMed] Article Kung, PP; Huang, B; Zehnder, L; Tatlock, J; Bingham, P; Krivacic, C; Gajiwala, K; Diehl, W; Yu, X; Maegley, KA SAH derived potent and selective EZH2 inhibitors. Bioorg Med Chem Lett25:1532-7 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone-lysine N-methyltransferase EZH1 |
|---|
| Name: | Histone-lysine N-methyltransferase EZH1 |
|---|
| Synonyms: | ENX-2 | EZH1 | EZH1_HUMAN | Enhancer of zeste homolog 1 | Enhancer of zeste homolog 1 (EZH1) | Histone-lysine N-methyltransferase EZH1 | KIAA0388 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 85285.34 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q92800 |
|---|
| Residue: | 747 |
|---|
| Sequence: | MEIPNPPTSKCITYWKRKVKSEYMRLRQLKRLQANMGAKALYVANFAKVQEKTQILNEEW
KKLRVQPVQSMKPVSGHPFLKKCTIESIFPGFASQHMLMRSLNTVALVPIMYSWSPLQQN
FMVEDETVLCNIPYMGDEVKEEDETFIEELINNYDGKVHGEEEMIPGSVLISDAVFLELV
DALNQYSDEEEEGHNDTSDGKQDDSKEDLPVTRKRKRHAIEGNKKSSKKQFPNDMIFSAI
ASMFPENGVPDDMKERYRELTEMSDPNALPPQCTPNIDGPNAKSVQREQSLHSFHTLFCR
RCFKYDCFLHPFHATPNVYKRKNKEIKIEPEPCGTDCFLLLEGAKEYAMLHNPRSKCSGR
RRRRHHIVSASCSNASASAVAETKEGDSDRDTGNDWASSSSEANSRCQTPTKQKASPAPP
QLCVVEAPSEPVEWTGAEESLFRVFHGTYFNNFCSIARLLGTKTCKQVFQFAVKESLILK
LPTDELMNPSQKKKRKHRLWAAHCRKIQLKKDNSSTQVYNYQPCDHPDRPCDSTCPCIMT
QNFCEKFCQCNPDCQNRFPGCRCKTQCNTKQCPCYLAVRECDPDLCLTCGASEHWDCKVV
SCKNCSIQRGLKKHLLLAPSDVAGWGTFIKESVQKNEFISEYCGELISQDEADRRGKVYD
KYMSSFLFNLNNDFVVDATRKGNKIRFANHSVNPNCYAKVVMVNGDHRIGIFAKRAIQAG
EELFFDYRYSQADALKYVGIERETDVL
|
|
|
|---|
| BDBM50009672 |
|---|
| n/a |
|---|
| Name | BDBM50009672 |
|---|
| Synonyms: | AdoHcy | CHEMBL418052 | S-(5'-adenosyl)-L-homocysteine | S-(5'-deoxyadenosin-5'-yl)-L-homocysteine | S-[1-(adenin-9-yl)-1,5-dideoxy-beta-D-ribofuranos-5-yl]-L-homocysteine | S-adenosyl-L-homocysteine | SAH | US8895245, S-Adenosyl-L-homocysteine (SAH) | US9175331, 1 | US9333217, S-Adenosyl-L-homocysteine (SAH) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H20N6O5S |
|---|
| Mol. Mass. | 384.411 |
|---|
| SMILES | N[C@@H](CCSC[C@H]1O[C@H]([C@H](O)[C@@H]1O)n1cnc2c(N)ncnc12)C(O)=O |
|---|
| Structure | 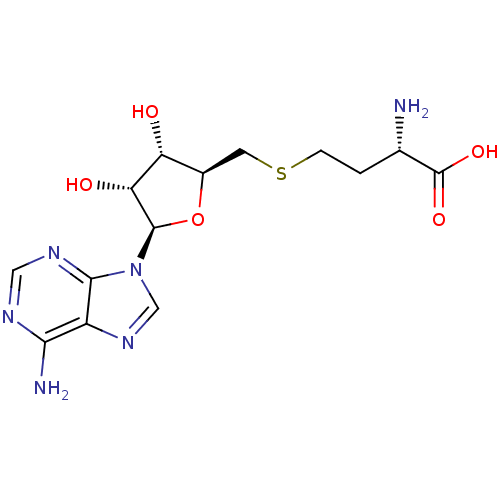
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kung, PP; Huang, B; Zehnder, L; Tatlock, J; Bingham, P; Krivacic, C; Gajiwala, K; Diehl, W; Yu, X; Maegley, KA SAH derived potent and selective EZH2 inhibitors. Bioorg Med Chem Lett25:1532-7 (2015) [PubMed] Article
Kung, PP; Huang, B; Zehnder, L; Tatlock, J; Bingham, P; Krivacic, C; Gajiwala, K; Diehl, W; Yu, X; Maegley, KA SAH derived potent and selective EZH2 inhibitors. Bioorg Med Chem Lett25:1532-7 (2015) [PubMed] Article