| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosine-protein kinase ZAP-70 |
|---|
| Ligand | BDBM50075735 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1473536 (CHEMBL3420415) |
|---|
| IC50 | 5.3±n/a nM |
|---|
| Citation |  Ellis, JM; Altman, MD; Bass, A; Butcher, JW; Byford, AJ; Donofrio, A; Galloway, S; Haidle, AM; Jewell, J; Kelly, N; Leccese, EK; Lee, S; Maddess, M; Miller, JR; Moy, LY; Osimboni, E; Otte, RD; Reddy, MV; Spencer, K; Sun, B; Vincent, SH; Ward, GJ; Woo, GH; Yang, C; Houshyar, H; Northrup, AB Overcoming mutagenicity and ion channel activity: optimization of selective spleen tyrosine kinase inhibitors. J Med Chem58:1929-39 (2015) [PubMed] Article Ellis, JM; Altman, MD; Bass, A; Butcher, JW; Byford, AJ; Donofrio, A; Galloway, S; Haidle, AM; Jewell, J; Kelly, N; Leccese, EK; Lee, S; Maddess, M; Miller, JR; Moy, LY; Osimboni, E; Otte, RD; Reddy, MV; Spencer, K; Sun, B; Vincent, SH; Ward, GJ; Woo, GH; Yang, C; Houshyar, H; Northrup, AB Overcoming mutagenicity and ion channel activity: optimization of selective spleen tyrosine kinase inhibitors. J Med Chem58:1929-39 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Tyrosine-protein kinase ZAP-70 |
|---|
| Name: | Tyrosine-protein kinase ZAP-70 |
|---|
| Synonyms: | 70 kDa zeta-associated protein | SRK | Syk-related tyrosine kinase | Tyrosine Kinase ZAP-70 | Tyrosine-protein kinase ZAP-70 | Tyrosine-protein kinase ZAP-70 (Syk) | Tyrosine-protein kinase ZAP-70 (ZAP70) | Tyrosine-protein kinase ZAP70 | ZAP70 | ZAP70_HUMAN | Zeta-chain (TCR) associated protein kinase 70kDa |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 69881.61 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ZAP-70 SH2 domain was expressed and purified from E. coli using O-phospho-L-tyrosine-agarose column. |
|---|
| Residue: | 619 |
|---|
| Sequence: | MPDPAAHLPFFYGSISRAEAEEHLKLAGMADGLFLLRQCLRSLGGYVLSLVHDVRFHHFP
IERQLNGTYAIAGGKAHCGPAELCEFYSRDPDGLPCNLRKPCNRPSGLEPQPGVFDCLRD
AMVRDYVRQTWKLEGEALEQAIISQAPQVEKLIATTAHERMPWYHSSLTREEAERKLYSG
AQTDGKFLLRPRKEQGTYALSLIYGKTVYHYLISQDKAGKYCIPEGTKFDTLWQLVEYLK
LKADGLIYCLKEACPNSSASNASGAAAPTLPAHPSTLTHPQRRIDTLNSDGYTPEPARIT
SPDKPRPMPMDTSVYESPYSDPEELKDKKLFLKRDNLLIADIELGCGNFGSVRQGVYRMR
KKQIDVAIKVLKQGTEKADTEEMMREAQIMHQLDNPYIVRLIGVCQAEALMLVMEMAGGG
PLHKFLVGKREEIPVSNVAELLHQVSMGMKYLEEKNFVHRDLAARNVLLVNRHYAKISDF
GLSKALGADDSYYTARSAGKWPLKWYAPECINFRKFSSRSDVWSYGVTMWEALSYGQKPY
KKMKGPEVMAFIEQGKRMECPPECPPELYALMSDCWIYKWEDRPDFLTVEQRMRACYYSL
ASKVEGPPGSTQKAEAACA
|
|
|
|---|
| BDBM50075735 |
|---|
| n/a |
|---|
| Name | BDBM50075735 |
|---|
| Synonyms: | CHEMBL3415583 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H26N6O |
|---|
| Mol. Mass. | 354.4493 |
|---|
| SMILES | Cc1cc(C)nc(Nc2cc(N[C@@H]3CCCC[C@@H]3N)cnc2C(N)=O)c1 |r| |
|---|
| Structure | 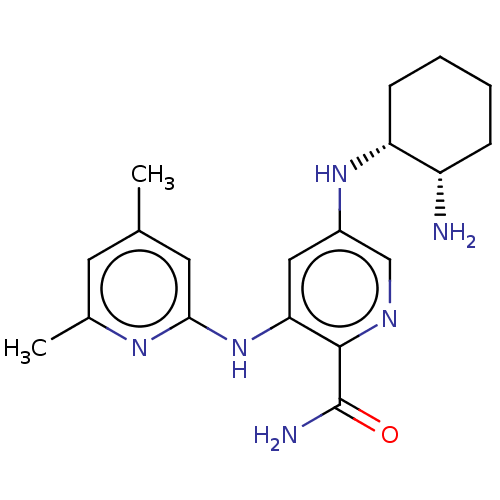
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ellis, JM; Altman, MD; Bass, A; Butcher, JW; Byford, AJ; Donofrio, A; Galloway, S; Haidle, AM; Jewell, J; Kelly, N; Leccese, EK; Lee, S; Maddess, M; Miller, JR; Moy, LY; Osimboni, E; Otte, RD; Reddy, MV; Spencer, K; Sun, B; Vincent, SH; Ward, GJ; Woo, GH; Yang, C; Houshyar, H; Northrup, AB Overcoming mutagenicity and ion channel activity: optimization of selective spleen tyrosine kinase inhibitors. J Med Chem58:1929-39 (2015) [PubMed] Article
Ellis, JM; Altman, MD; Bass, A; Butcher, JW; Byford, AJ; Donofrio, A; Galloway, S; Haidle, AM; Jewell, J; Kelly, N; Leccese, EK; Lee, S; Maddess, M; Miller, JR; Moy, LY; Osimboni, E; Otte, RD; Reddy, MV; Spencer, K; Sun, B; Vincent, SH; Ward, GJ; Woo, GH; Yang, C; Houshyar, H; Northrup, AB Overcoming mutagenicity and ion channel activity: optimization of selective spleen tyrosine kinase inhibitors. J Med Chem58:1929-39 (2015) [PubMed] Article