| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 3-phosphoinositide-dependent protein kinase 1 |
|---|
| Ligand | BDBM50078207 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1470575 (CHEMBL3419801) |
|---|
| Kd | 11100±n/a nM |
|---|
| Citation |  Yan, R; Chuang, HC; Kapuriya, N; Chou, CC; Lai, PT; Chang, HW; Yang, CN; Kulp, SK; Chen, CS Exploitation of the ability of¿-tocopherol to facilitate membrane co-localization of Akt and PHLPP1 to develop PHLPP1-targeted Akt inhibitors. J Med Chem58:2290-8 (2015) [PubMed] Article Yan, R; Chuang, HC; Kapuriya, N; Chou, CC; Lai, PT; Chang, HW; Yang, CN; Kulp, SK; Chen, CS Exploitation of the ability of¿-tocopherol to facilitate membrane co-localization of Akt and PHLPP1 to develop PHLPP1-targeted Akt inhibitors. J Med Chem58:2290-8 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 3-phosphoinositide-dependent protein kinase 1 |
|---|
| Name: | 3-phosphoinositide-dependent protein kinase 1 |
|---|
| Synonyms: | 3-Phosphoinositide-Dependent Protein Kinase 1 (PDK1) | 3-phosphoinositide dependent protein kinase-1 | 3-phosphoinositide-dependent protein kinase 1 | 3-phosphoinositide-dependent protein kinase 1 (PDK) | 3-phosphoinositide-dependent protein kinase 1 (PDK-1) | 3-phosphoinositide-dependent protein kinase 1 (PDK1)(Δ1-50) | Isoform 2 of 3-phosphoinositide-dependent protein kinase 1 | PDK1 | PDPK1 | PDPK1_HUMAN | Phosphoinositide-dependent protein kinase 1 (PDK1) | Pyruvate dehydrogenase kinase isoenzyme 1 (PDK1) | hPDK1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 63157.65 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15530 |
|---|
| Residue: | 556 |
|---|
| Sequence: | MARTTSQLYDAVPIQSSVVLCSCPSPSMVRTQTESSTPPGIPGGSRQGPAMDGTAAEPRP
GAGSLQHAQPPPQPRKKRPEDFKFGKILGEGSFSTVVLARELATSREYAIKILEKRHIIK
ENKVPYVTRERDVMSRLDHPFFVKLYFTFQDDEKLYFGLSYAKNGELLKYIRKIGSFDET
CTRFYTAEIVSALEYLHGKGIIHRDLKPENILLNEDMHIQITDFGTAKVLSPESKQARAN
SFVGTAQYVSPELLTEKSACKSSDLWALGCIIYQLVAGLPPFRAGNEYLIFQKIIKLEYD
FPEKFFPKARDLVEKLLVLDATKRLGCEEMEGYGPLKAHPFFESVTWENLHQQTPPKLTA
YLPAMSEDDEDCYGNYDNLLSQFGCMQVSSSSSSHSLSASDTGLPQRSGSNIEQYIHDLD
SNSFELDLQFSEDEKRLLLEKQAGGNPWHQFVENNLILKMGPVDKRKGLFARRRQLLLTE
GPHLYYVDPVNKVLKGEIPWSQELRPEAKNFKTFFVHTPNRTYYLMDPSGNAHKWCRKIQ
EVWRQRYQSHPDAAVQ
|
|
|
|---|
| BDBM50078207 |
|---|
| n/a |
|---|
| Name | BDBM50078207 |
|---|
| Synonyms: | CHEMBL3414688 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H24BrNO3S |
|---|
| Mol. Mass. | 390.336 |
|---|
| SMILES | CC(C)CCCC1(C)CCc2cc(cc(Br)c2O1)S(N)(=O)=O |
|---|
| Structure | 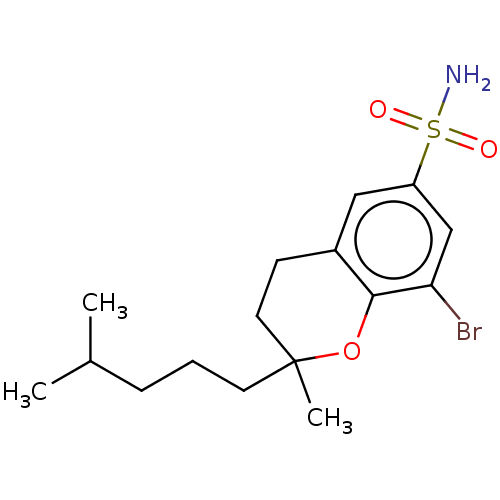
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Yan, R; Chuang, HC; Kapuriya, N; Chou, CC; Lai, PT; Chang, HW; Yang, CN; Kulp, SK; Chen, CS Exploitation of the ability of¿-tocopherol to facilitate membrane co-localization of Akt and PHLPP1 to develop PHLPP1-targeted Akt inhibitors. J Med Chem58:2290-8 (2015) [PubMed] Article
Yan, R; Chuang, HC; Kapuriya, N; Chou, CC; Lai, PT; Chang, HW; Yang, CN; Kulp, SK; Chen, CS Exploitation of the ability of¿-tocopherol to facilitate membrane co-localization of Akt and PHLPP1 to develop PHLPP1-targeted Akt inhibitors. J Med Chem58:2290-8 (2015) [PubMed] Article