

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 | ||
| Ligand | BDBM50086319 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1476004 (CHEMBL3428360) | ||
| IC50 | 9.0±n/a nM | ||
| Citation |  Haffner, CD; Becherer, JD; Boros, EE; Cadilla, R; Carpenter, T; Cowan, D; Deaton, DN; Guo, Y; Harrington, W; Henke, BR; Jeune, MR; Kaldor, I; Milliken, N; Petrov, KG; Preugschat, F; Schulte, C; Shearer, BG; Shearer, T; Smalley, TL; Stewart, EL; Stuart, JD; Ulrich, JC Discovery, Synthesis, and Biological Evaluation of Thiazoloquin(az)olin(on)es as Potent CD38 Inhibitors. J Med Chem58:3548-71 (2015) [PubMed] Article Haffner, CD; Becherer, JD; Boros, EE; Cadilla, R; Carpenter, T; Cowan, D; Deaton, DN; Guo, Y; Harrington, W; Henke, BR; Jeune, MR; Kaldor, I; Milliken, N; Petrov, KG; Preugschat, F; Schulte, C; Shearer, BG; Shearer, T; Smalley, TL; Stewart, EL; Stuart, JD; Ulrich, JC Discovery, Synthesis, and Biological Evaluation of Thiazoloquin(az)olin(on)es as Potent CD38 Inhibitors. J Med Chem58:3548-71 (2015) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 | |||
| Name: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 | ||
| Synonyms: | ADP-ribosyl cyclase 1 | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 | ADPRC 1 | CD38_MOUSE | CD_antigen=CD38 | Cd38 | Cyclic ADP-ribose hydrolase 1 | I-19 | NIM-R5 antigen | cADPr hydrolase 1 | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 34417.04 | ||
| Organism: | Mus musculus | ||
| Description: | ChEMBL_109609 | ||
| Residue: | 304 | ||
| Sequence: |
| ||
| BDBM50086319 | |||
| n/a | |||
| Name | BDBM50086319 | ||
| Synonyms: | CHEMBL3426191 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C18H19N5OS | ||
| Mol. Mass. | 353.441 | ||
| SMILES | NC(=O)[C@H]1CC[C@@H](CC1)Nc1ncnc2ccc(cc12)-c1cncs1 |r,wU:3.2,wD:6.9,(6.69,7.37,;6.68,6.14,;7.75,5.52,;5.34,5.38,;5.34,3.84,;4,3.07,;2.67,3.85,;2.68,5.39,;4.01,6.15,;1.33,3.08,;1.33,1.54,;2.66,.77,;2.66,-.77,;1.33,-1.54,;,-.77,;-1.33,-1.54,;-2.68,-.77,;-2.68,.77,;-1.33,1.54,;,.77,;-4.01,1.54,;-5.4,.91,;-6.43,2.07,;-5.65,3.39,;-4.14,3.06,)| | ||
| Structure | 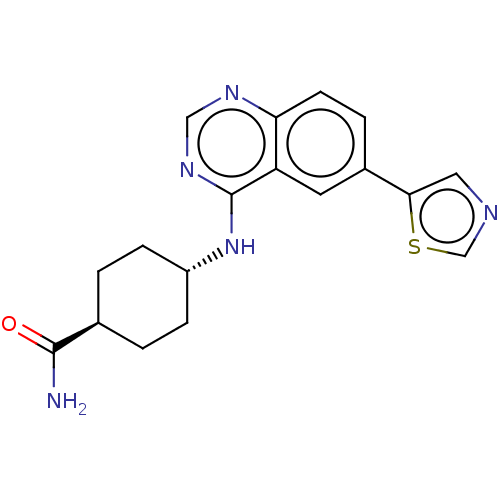
| ||