

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Induced myeloid leukemia cell differentiation protein Mcl-1 | ||
| Ligand | BDBM50086706 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1477334 (CHEMBL3429300) | ||
| Ki | 0.454000±n/a nM | ||
| Citation |  Burke, JP; Bian, Z; Shaw, S; Zhao, B; Goodwin, CM; Belmar, J; Browning, CF; Vigil, D; Friberg, A; Camper, DV; Rossanese, OW; Lee, T; Olejniczak, ET; Fesik, SW Discovery of tricyclic indoles that potently inhibit Mcl-1 using fragment-based methods and structure-based design. J Med Chem58:3794-805 (2015) [PubMed] Article Burke, JP; Bian, Z; Shaw, S; Zhao, B; Goodwin, CM; Belmar, J; Browning, CF; Vigil, D; Friberg, A; Camper, DV; Rossanese, OW; Lee, T; Olejniczak, ET; Fesik, SW Discovery of tricyclic indoles that potently inhibit Mcl-1 using fragment-based methods and structure-based design. J Med Chem58:3794-805 (2015) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Induced myeloid leukemia cell differentiation protein Mcl-1 | |||
| Name: | Induced myeloid leukemia cell differentiation protein Mcl-1 | ||
| Synonyms: | BCL2L3 | Bcl-2-like protein 3 | Bcl-2-like protein 3 (Mcl-1) | Bcl-2-related protein EAT/mcl1 | Bcl2-L-3 | Induced myeloid leukemia cell differentiation protein (Mcl-1) | MCL1 | MCL1_HUMAN | Mcl-1 | Myeloid Cell factor-1 (Mcl-1) | Myeloid cell leukemia sequence 1 (BCL2-related) | mcl1/EAT | ||
| Type: | Membrane; Single-pass membrane protein | ||
| Mol. Mass.: | 37332.87 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q07820 | ||
| Residue: | 350 | ||
| Sequence: |
| ||
| BDBM50086706 | |||
| n/a | |||
| Name | BDBM50086706 | ||
| Synonyms: | CHEMBL3426335 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C37H51N7O7S | ||
| Mol. Mass. | 737.909 | ||
| SMILES | COCCCc1c(C(O)=O)n(CCN2CCOCC2)c2c(cccc12)-c1c(C)nn(C)c1COc1ccc(cc1)N1CCN(CC1)S(=O)(=O)N(C)C |(6.12,5.96,;5.74,4.79,;4.23,4.48,;3.75,3.01,;2.24,2.7,;1.76,1.24,;2.66,.02,;4.2,.04,;4.83,-1.02,;4.8,1.11,;1.76,-1.24,;2.24,-2.7,;3.75,-3.01,;4.24,-4.47,;5.75,-4.78,;6.23,-6.25,;5.2,-7.4,;3.69,-7.08,;3.21,-5.62,;.3,-.77,;-1.03,-1.55,;-2.38,-.77,;-2.38,.77,;-1.03,1.55,;.3,.77,;-1.02,-3.09,;.23,-3.96,;1.4,-3.56,;-.23,-5.43,;-1.77,-5.45,;-2.48,-6.45,;-2.26,-3.99,;-3.73,-3.52,;-4.87,-4.56,;-6.33,-4.09,;-7.48,-5.12,;-8.94,-4.65,;-9.27,-3.14,;-8.12,-2.11,;-6.66,-2.58,;-10.73,-2.67,;-11.88,-3.7,;-13.34,-3.23,;-13.66,-1.72,;-12.52,-.69,;-11.05,-1.16,;-15.13,-1.25,;-15.39,-.04,;-16.3,-.87,;-16.27,-2.28,;-17.45,-1.9,;-16.02,-3.48,)| | ||
| Structure | 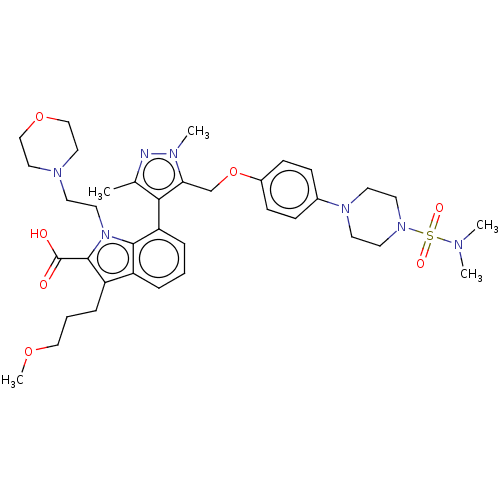
| ||