| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glutamate carboxypeptidase 2 |
|---|
| Ligand | BDBM50392045 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1487181 (CHEMBL3533357) |
|---|
| Ki | 1.000000±n/a nM |
|---|
| Citation |  Rais, R; Hoover, R; Wozniak, K; Rudek, MA; Tsukamoto, T; Alt, J; Rojas, C; Slusher, BS Reversible disulfide formation of the glutamate carboxypeptidase II inhibitor E2072 results in prolonged systemic exposures in vivo. Drug Metab Dispos40:2315-23 (2012) [PubMed] Article Rais, R; Hoover, R; Wozniak, K; Rudek, MA; Tsukamoto, T; Alt, J; Rojas, C; Slusher, BS Reversible disulfide formation of the glutamate carboxypeptidase II inhibitor E2072 results in prolonged systemic exposures in vivo. Drug Metab Dispos40:2315-23 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Glutamate carboxypeptidase 2 |
|---|
| Name: | Glutamate carboxypeptidase 2 |
|---|
| Synonyms: | FGCP | FOLH | FOLH1 | FOLH1_HUMAN | Folate hydrolase 1 | Folylpoly-gamma-glutamate carboxypeptidase | Glutamate carboxypeptidase 2 | Glutamate carboxypeptidase II | Membrane glutamate carboxypeptidase | N-acetylated-alpha-linked acidic dipeptidase I | NAALAD1 | NAALADase I | PSM | PSMA | Prostate-specific membrane antigen | Pteroylpoly-gamma-glutamate carboxypeptidase | mGCP |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 84333.66 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1497035 |
|---|
| Residue: | 750 |
|---|
| Sequence: | MWNLLHETDSAVATARRPRWLCAGALVLAGGFFLLGFLFGWFIKSSNEATNITPKHNMKA
FLDELKAENIKKFLYNFTQIPHLAGTEQNFQLAKQIQSQWKEFGLDSVELAHYDVLLSYP
NKTHPNYISIINEDGNEIFNTSLFEPPPPGYENVSDIVPPFSAFSPQGMPEGDLVYVNYA
RTEDFFKLERDMKINCSGKIVIARYGKVFRGNKVKNAQLAGAKGVILYSDPADYFAPGVK
SYPDGWNLPGGGVQRGNILNLNGAGDPLTPGYPANEYAYRRGIAEAVGLPSIPVHPIGYY
DAQKLLEKMGGSAPPDSSWRGSLKVPYNVGPGFTGNFSTQKVKMHIHSTNEVTRIYNVIG
TLRGAVEPDRYVILGGHRDSWVFGGIDPQSGAAVVHEIVRSFGTLKKEGWRPRRTILFAS
WDAEEFGLLGSTEWAEENSRLLQERGVAYINADSSIEGNYTLRVDCTPLMYSLVHNLTKE
LKSPDEGFEGKSLYESWTKKSPSPEFSGMPRISKLGSGNDFEVFFQRLGIASGRARYTKN
WETNKFSGYPLYHSVYETYELVEKFYDPMFKYHLTVAQVRGGMVFELANSIVLPFDCRDY
AVVLRKYADKIYSISMKHPQEMKTYSVSFDSLFSAVKNFTEIASKFSERLQDFDKSNPIV
LRMMNDQLMFLERAFIDPLGLPDRPFYRHVIYAPSSHNKYAGESFPGIYDALFDIESKVD
PSKAWGEVKRQIYVAAFTVQAAAETLSEVA
|
|
|
|---|
| BDBM50392045 |
|---|
| n/a |
|---|
| Name | BDBM50392045 |
|---|
| Synonyms: | CHEMBL2152561 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H14O4S |
|---|
| Mol. Mass. | 302.345 |
|---|
| SMILES | OC(=O)c1cccc(c1)-c1cccc(CCS)c1C(O)=O |
|---|
| Structure | 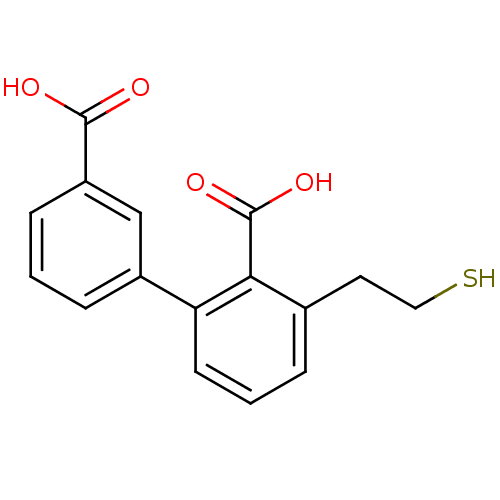
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Rais, R; Hoover, R; Wozniak, K; Rudek, MA; Tsukamoto, T; Alt, J; Rojas, C; Slusher, BS Reversible disulfide formation of the glutamate carboxypeptidase II inhibitor E2072 results in prolonged systemic exposures in vivo. Drug Metab Dispos40:2315-23 (2012) [PubMed] Article
Rais, R; Hoover, R; Wozniak, K; Rudek, MA; Tsukamoto, T; Alt, J; Rojas, C; Slusher, BS Reversible disulfide formation of the glutamate carboxypeptidase II inhibitor E2072 results in prolonged systemic exposures in vivo. Drug Metab Dispos40:2315-23 (2012) [PubMed] Article