| Reaction Details |
|---|
 | Report a problem with these data |
| Target | UDP-glucuronosyltransferase 1A8 |
|---|
| Ligand | BDBM50358252 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1486245 (CHEMBL3534364) |
|---|
| Ki | 354000±n/a nM |
|---|
| Citation |  Shiraga, T; Yajima, K; Suzuki, K; Suzuki, K; Hashimoto, T; Iwatsubo, T; Miyashita, A; Usui, T Identification of UDP-glucuronosyltransferases responsible for the glucuronidation of darexaban, an oral factor Xa inhibitor, in human liver and intestine. Drug Metab Dispos40:276-82 (2012) [PubMed] Article Shiraga, T; Yajima, K; Suzuki, K; Suzuki, K; Hashimoto, T; Iwatsubo, T; Miyashita, A; Usui, T Identification of UDP-glucuronosyltransferases responsible for the glucuronidation of darexaban, an oral factor Xa inhibitor, in human liver and intestine. Drug Metab Dispos40:276-82 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| UDP-glucuronosyltransferase 1A8 |
|---|
| Name: | UDP-glucuronosyltransferase 1A8 |
|---|
| Synonyms: | GNT1 | GNT1 | UD18_HUMAN | UDP-glucuronosyltransferase 1-8 | UDP-glucuronosyltransferase 1-H | UDP-glucuronosyltransferase 1A8 | UDPGT 1-8 | UGT-1H | UGT1 | UGT1*8 | UGT1-08 | UGT1.8 | UGT1A8 | UGT1H |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 59751.98 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_104086 |
|---|
| Residue: | 530 |
|---|
| Sequence: | MARTGWTSPIPLCVSLLLTCGFAEAGKLLVVPMDGSHWFTMQSVVEKLILRGHEVVVVMP
EVSWQLGKSLNCTVKTYSTSYTLEDLDREFMDFADAQWKAQVRSLFSLFLSSSNGFFNLF
FSHCRSLFNDRKLVEYLKESSFDAVFLDPFDACGLIVAKYFSLPSVVFARGIACHYLEEG
AQCPAPLSYVPRILLGFSDAMTFKERVRNHIMHLEEHLFCQYFSKNALEIASEILQTPVT
AYDLYSHTSIWLLRTDFVLDYPKPVMPNMIFIGGINCHQGKPLPMEFEAYINASGEHGIV
VFSLGSMVSEIPEKKAMAIADALGKIPQTVLWRYTGTRPSNLANNTILVKWLPQNDLLGH
PMTRAFITHAGSHGVYESICNGVPMVMMPLFGDQMDNAKRMETKGAGVTLNVLEMTSEDL
ENALKAVINDKSYKENIMRLSSLHKDRPVEPLDLAVFWVEFVMRHKGAPHLRPAAHDLTW
YQYHSLDVIGFLLAVVLTVAFITFKCCAYGYRKCLGKKGRVKKAHKSKTH
|
|
|
|---|
| BDBM50358252 |
|---|
| n/a |
|---|
| Name | BDBM50358252 |
|---|
| Synonyms: | CHEMBL1922235 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H30N4O4 |
|---|
| Mol. Mass. | 474.5515 |
|---|
| SMILES | COc1ccc(cc1)C(=O)Nc1cccc(O)c1NC(=O)c1ccc(cc1)N1CCCN(C)CC1 |
|---|
| Structure | 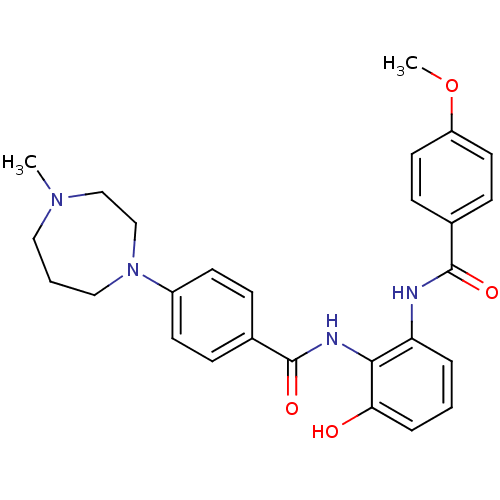
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Shiraga, T; Yajima, K; Suzuki, K; Suzuki, K; Hashimoto, T; Iwatsubo, T; Miyashita, A; Usui, T Identification of UDP-glucuronosyltransferases responsible for the glucuronidation of darexaban, an oral factor Xa inhibitor, in human liver and intestine. Drug Metab Dispos40:276-82 (2012) [PubMed] Article
Shiraga, T; Yajima, K; Suzuki, K; Suzuki, K; Hashimoto, T; Iwatsubo, T; Miyashita, A; Usui, T Identification of UDP-glucuronosyltransferases responsible for the glucuronidation of darexaban, an oral factor Xa inhibitor, in human liver and intestine. Drug Metab Dispos40:276-82 (2012) [PubMed] Article