

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Mu-type opioid receptor | ||
| Ligand | BDBM50090755 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1498300 (CHEMBL3583923) | ||
| EC50 | 0.220000±n/a nM | ||
| Citation |  Cueva, JP; Roche, C; Ostovar, M; Kumar, V; Clark, MJ; Hillhouse, TM; Lewis, JW; Traynor, JR; Husbands, SM C7ß-methyl analogues of the orvinols: the discovery of kappa opioid antagonists with nociceptin/orphanin FQ peptide (NOP) receptor partial agonism and low, or zero, efficacy at mu opioid receptors. J Med Chem58:4242-9 (2015) [PubMed] Article Cueva, JP; Roche, C; Ostovar, M; Kumar, V; Clark, MJ; Hillhouse, TM; Lewis, JW; Traynor, JR; Husbands, SM C7ß-methyl analogues of the orvinols: the discovery of kappa opioid antagonists with nociceptin/orphanin FQ peptide (NOP) receptor partial agonism and low, or zero, efficacy at mu opioid receptors. J Med Chem58:4242-9 (2015) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Mu-type opioid receptor | |||
| Name: | Mu-type opioid receptor | ||
| Synonyms: | MOR-1 | MUOR1 | Mu-type opioid receptor (MOR) | OPIATE Mu | OPRM_RAT | Opiate non-selective | Opioid receptor B | Oprm1 | Ror-b | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 44503.11 | ||
| Organism: | Rattus norvegicus (rat) | ||
| Description: | Competition binding assays were carried out using membrane preparations from transfected HN9.10 cells that constitutively expressed the mu opioid receptor. | ||
| Residue: | 398 | ||
| Sequence: |
| ||
| BDBM50090755 | |||
| n/a | |||
| Name | BDBM50090755 | ||
| Synonyms: | CHEMBL3581743 | US9259422, 22, R = 4-FPh- BU10120 | US9480684, 22, R = 4- FPh- BU10120 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C31H34FNO4 | ||
| Mol. Mass. | 503.6044 | ||
| SMILES | [H][C@@]12Oc3c4c(C[C@@]5([H])N(CC6CC6)CC[C@@]14[C@]51C[C@](C)([C@H](O)c4ccc(F)cc4)[C@]2(OC)C=C1)ccc3O |r,wU:17.21,16.16,19.23,1.0,wD:30.36,21.25,7.7,c:39,TLB:21:19:16.1:34.33,THB:10:9:17:4.5.6,20:19:16.1:34.33,(-3.17,.26,;-2.2,.57,;-3.38,2.21,;-2.2,3.8,;-.81,2.99,;.55,3.8,;1.94,2.99,;1.94,1.4,;2.83,1.9,;3.33,.57,;4.33,-.58,;5.84,-.3,;7.2,-.82,;6.92,.69,;2.73,2.15,;.51,2.18,;-.81,1.4,;.55,.57,;.55,-.99,;-.81,-1.79,;-1.89,-2.38,;.53,-2.51,;1.56,-1.84,;.61,-4.05,;-.68,-4.89,;-.6,-6.42,;.77,-7.12,;.84,-8.35,;2.06,-6.28,;1.98,-4.74,;-2.2,-.99,;-3.6,-1.56,;-4.57,-.8,;-.88,-.72,;-.88,.28,;.55,5.4,;-.81,6.2,;-2.2,5.4,;-3.26,6.01,)| | ||
| Structure | 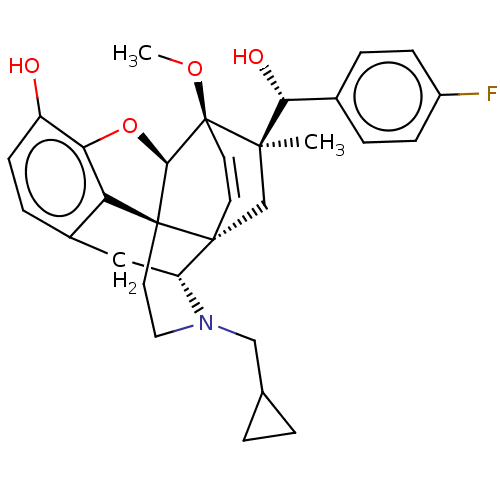
| ||