| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fibroblast growth factor receptor 4 |
|---|
| Ligand | BDBM50092080 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1498711 (CHEMBL3583124) |
|---|
| IC50 | 5900±n/a nM |
|---|
| Citation |  Choi, HS; Rucker, PV; Wang, Z; Fan, Y; Albaugh, P; Chopiuk, G; Gessier, F; Sun, F; Adrian, F; Liu, G; Hood, T; Li, N; Jia, Y; Che, J; McCormack, S; Li, A; Li, J; Steffy, A; Culazzo, A; Tompkins, C; Phung, V; Kreusch, A; Lu, M; Hu, B; Chaudhary, A; Prashad, M; Tuntland, T; Liu, B; Harris, J; Seidel, HM; Loren, J; Molteni, V (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors. ACS Med Chem Lett6:562-7 (2015) [PubMed] Article Choi, HS; Rucker, PV; Wang, Z; Fan, Y; Albaugh, P; Chopiuk, G; Gessier, F; Sun, F; Adrian, F; Liu, G; Hood, T; Li, N; Jia, Y; Che, J; McCormack, S; Li, A; Li, J; Steffy, A; Culazzo, A; Tompkins, C; Phung, V; Kreusch, A; Lu, M; Hu, B; Chaudhary, A; Prashad, M; Tuntland, T; Liu, B; Harris, J; Seidel, HM; Loren, J; Molteni, V (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors. ACS Med Chem Lett6:562-7 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fibroblast growth factor receptor 4 |
|---|
| Name: | Fibroblast growth factor receptor 4 |
|---|
| Synonyms: | CD_antigen: CD334 FGFR4 | FGFR-4 | FGFR4 | FGFR4_HUMAN | Fibroblast growth factor receptor | Fibroblast growth factor receptor 4 (FGFR4) | JK2 | JTK2 | TKF |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 87957.45 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P22455 |
|---|
| Residue: | 802 |
|---|
| Sequence: | MRLLLALLGVLLSVPGPPVLSLEASEEVELEPCLAPSLEQQEQELTVALGQPVRLCCGRA
ERGGHWYKEGSRLAPAGRVRGWRGRLEIASFLPEDAGRYLCLARGSMIVLQNLTLITGDS
LTSSNDDEDPKSHRDPSNRHSYPQQAPYWTHPQRMEKKLHAVPAGNTVKFRCPAAGNPTP
TIRWLKDGQAFHGENRIGGIRLRHQHWSLVMESVVPSDRGTYTCLVENAVGSIRYNYLLD
VLERSPHRPILQAGLPANTTAVVGSDVELLCKVYSDAQPHIQWLKHIVINGSSFGADGFP
YVQVLKTADINSSEVEVLYLRNVSAEDAGEYTCLAGNSIGLSYQSAWLTVLPEEDPTWTA
AAPEARYTDIILYASGSLALAVLLLLAGLYRGQALHGRHPRPPATVQKLSRFPLARQFSL
ESGSSGKSSSSLVRGVRLSSSGPALLAGLVSLDLPLDPLWEFPRDRLVLGKPLGEGCFGQ
VVRAEAFGMDPARPDQASTVAVKMLKDNASDKDLADLVSEMEVMKLIGRHKNIINLLGVC
TQEGPLYVIVECAAKGNLREFLRARRPPGPDLSPDGPRSSEGPLSFPVLVSCAYQVARGM
QYLESRKCIHRDLAARNVLVTEDNVMKIADFGLARGVHHIDYYKKTSNGRLPVKWMAPEA
LFDRVYTHQSDVWSFGILLWEIFTLGGSPYPGIPVEELFSLLREGHRMDRPPHCPPELYG
LMRECWHAAPSQRPTFKQLVEALDKVLLAVSEEYLDLRLTFGPYSPSGGDASSTCSSSDS
VFSHDPLPLGSSSFPFGSGVQT
|
|
|
|---|
| BDBM50092080 |
|---|
| n/a |
|---|
| Name | BDBM50092080 |
|---|
| Synonyms: | CHEMBL3582427 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H15N5 |
|---|
| Mol. Mass. | 325.3666 |
|---|
| SMILES | N#Cc1ccc(cc1)-c1cnc2ccc(NCc3ccccc3)nn12 |
|---|
| Structure | 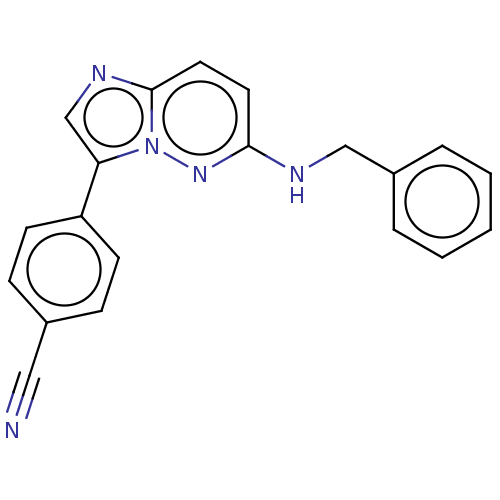
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Choi, HS; Rucker, PV; Wang, Z; Fan, Y; Albaugh, P; Chopiuk, G; Gessier, F; Sun, F; Adrian, F; Liu, G; Hood, T; Li, N; Jia, Y; Che, J; McCormack, S; Li, A; Li, J; Steffy, A; Culazzo, A; Tompkins, C; Phung, V; Kreusch, A; Lu, M; Hu, B; Chaudhary, A; Prashad, M; Tuntland, T; Liu, B; Harris, J; Seidel, HM; Loren, J; Molteni, V (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors. ACS Med Chem Lett6:562-7 (2015) [PubMed] Article
Choi, HS; Rucker, PV; Wang, Z; Fan, Y; Albaugh, P; Chopiuk, G; Gessier, F; Sun, F; Adrian, F; Liu, G; Hood, T; Li, N; Jia, Y; Che, J; McCormack, S; Li, A; Li, J; Steffy, A; Culazzo, A; Tompkins, C; Phung, V; Kreusch, A; Lu, M; Hu, B; Chaudhary, A; Prashad, M; Tuntland, T; Liu, B; Harris, J; Seidel, HM; Loren, J; Molteni, V (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors. ACS Med Chem Lett6:562-7 (2015) [PubMed] Article