

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Bifunctional epoxide hydrolase 2 | ||
| Ligand | BDBM50100531 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1455803 (CHEMBL3361515) | ||
| Ki | 0.230000±n/a nM | ||
| Citation |  Lee, KS; Liu, JY; Wagner, KM; Pakhomova, S; Dong, H; Morisseau, C; Fu, SH; Yang, J; Wang, P; Ulu, A; Mate, CA; Nguyen, LV; Hwang, SH; Edin, ML; Mara, AA; Wulff, H; Newcomer, ME; Zeldin, DC; Hammock, BD Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy. J Med Chem57:7016-30 (2014) [PubMed] Article Lee, KS; Liu, JY; Wagner, KM; Pakhomova, S; Dong, H; Morisseau, C; Fu, SH; Yang, J; Wang, P; Ulu, A; Mate, CA; Nguyen, LV; Hwang, SH; Edin, ML; Mara, AA; Wulff, H; Newcomer, ME; Zeldin, DC; Hammock, BD Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy. J Med Chem57:7016-30 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Bifunctional epoxide hydrolase 2 | |||
| Name: | Bifunctional epoxide hydrolase 2 | ||
| Synonyms: | Cytosolic epoxide hydrolase 2 | EBifunctional epoxide hydrolase 2 | EPHX2 | Epoxide hydratase | HYES_HUMAN | Lipid-phosphate phosphatase | Soluble epoxide hydrolase (sEH) | epoxide hydrolase 2, cytoplasmic | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 62613.07 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P34913 | ||
| Residue: | 555 | ||
| Sequence: |
| ||
| BDBM50100531 | |||
| n/a | |||
| Name | BDBM50100531 | ||
| Synonyms: | CHEMBL3325465 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C19H35N3O2 | ||
| Mol. Mass. | 337.5001 | ||
| SMILES | CCC(C)C(=O)N1CCC(CC1)NC(=O)N[C@H]1CC[C@H](CC)CC1 |r,wU:16.16,wD:19.20,(21.72,-50.53,;20.39,-49.76,;20.39,-48.22,;19.05,-47.45,;21.72,-47.45,;21.72,-45.91,;23.06,-48.22,;24.38,-47.45,;25.72,-48.22,;25.72,-49.76,;24.38,-50.53,;23.06,-49.76,;27.06,-50.53,;28.39,-49.76,;29.72,-50.53,;28.39,-48.22,;29.72,-47.45,;29.72,-45.91,;31.06,-45.14,;32.39,-45.91,;33.72,-45.14,;35.06,-45.91,;32.39,-47.45,;31.06,-48.22,)| | ||
| Structure | 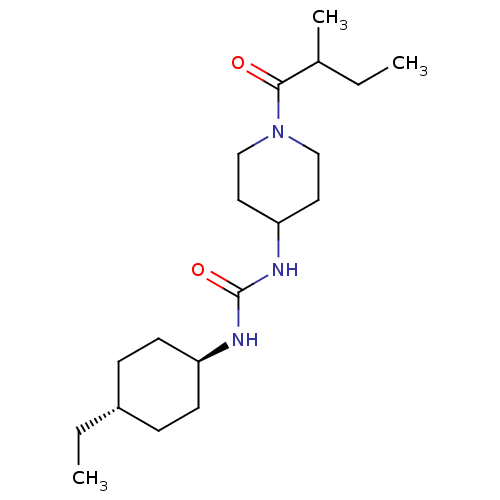
| ||