| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Polyamine deacetylase HDAC10 |
|---|
| Ligand | BDBM50101330 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1455103 (CHEMBL3366359) |
|---|
| IC50 | 0.500000±n/a nM |
|---|
| Citation |  Salvador, LA; Park, H; Al-Awadhi, FH; Liu, Y; Kim, B; Zeller, SL; Chen, QY; Hong, J; Luesch, H Modulation of Activity Profiles for Largazole-Based HDAC Inhibitors through Alteration of Prodrug Properties. ACS Med Chem Lett5:905-10 (2014) [PubMed] Article Salvador, LA; Park, H; Al-Awadhi, FH; Liu, Y; Kim, B; Zeller, SL; Chen, QY; Hong, J; Luesch, H Modulation of Activity Profiles for Largazole-Based HDAC Inhibitors through Alteration of Prodrug Properties. ACS Med Chem Lett5:905-10 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Polyamine deacetylase HDAC10 |
|---|
| Name: | Polyamine deacetylase HDAC10 |
|---|
| Synonyms: | HD10 | HDA10_HUMAN | HDAC10 | Histone deacetylase | Histone deacetylase 10 | Human HDAC10 |
|---|
| Type: | Chromatin regulator; hydrolase; repressor |
|---|
| Mol. Mass.: | 71431.89 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q969S8 |
|---|
| Residue: | 669 |
|---|
| Sequence: | MGTALVYHEDMTATRLLWDDPECEIERPERLTAALDRLRQRGLEQRCLRLSAREASEEEL
GLVHSPEYVSLVRETQVLGKEELQALSGQFDAIYFHPSTFHCARLAAGAGLQLVDAVLTG
AVQNGLALVRPPGHHGQRAAANGFCVFNNVAIAAAHAKQKHGLHRILVVDWDVHHGQGIQ
YLFEDDPSVLYFSWHRYEHGRFWPFLRESDADAVGRGQGLGFTVNLPWNQVGMGNADYVA
AFLHLLLPLAFEFDPELVLVSAGFDSAIGDPEGQMQATPECFAHLTQLLQVLAGGRVCAV
LEGGYHLESLAESVCMTVQTLLGDPAPPLSGPMAPCQSALESIQSARAAQAPHWKSLQQQ
DVTAVPMSPSSHSPEGRPPPLLPGGPVCKAAASAPSSLLDQPCLCPAPSVRTAVALTTPD
ITLVLPPDVIQQEASALREETEAWARPHESLAREEALTALGKLLYLLDGMLDGQVNSGIA
ATPASAAAATLDVAVRRGLSHGAQRLLCVALGQLDRPPDLAHDGRSLWLNIRGKEAAALS
MFHVSTPLPVMTGGFLSCILGLVLPLAYGFQPDLVLVALGPGHGLQGPHAALLAAMLRGL
AGGRVLALLEENSTPQLAGILARVLNGEAPPSLGPSSVASPEDVQALMYLRGQLEPQWKM
LQCHPHLVA
|
|
|
|---|
| BDBM50101330 |
|---|
| n/a |
|---|
| Name | BDBM50101330 |
|---|
| Synonyms: | CHEMBL3329622 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C33H49N5O8S4 |
|---|
| Mol. Mass. | 772.031 |
|---|
| SMILES | CC(C)[C@@H]1NC(=O)[C@]2(C)CSC(=N2)c2csc(CNC(=O)C[C@H](OC1=O)\C=C\CCSSC[C@H](NC(=O)OC(C)(C)C)C(=O)OC(C)(C)C)n2 |r,c:11| |
|---|
| Structure | 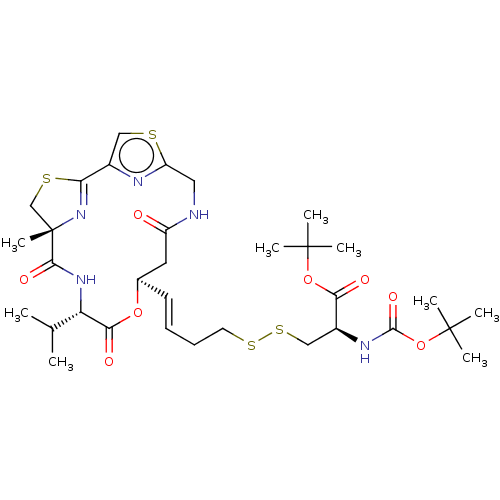
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Salvador, LA; Park, H; Al-Awadhi, FH; Liu, Y; Kim, B; Zeller, SL; Chen, QY; Hong, J; Luesch, H Modulation of Activity Profiles for Largazole-Based HDAC Inhibitors through Alteration of Prodrug Properties. ACS Med Chem Lett5:905-10 (2014) [PubMed] Article
Salvador, LA; Park, H; Al-Awadhi, FH; Liu, Y; Kim, B; Zeller, SL; Chen, QY; Hong, J; Luesch, H Modulation of Activity Profiles for Largazole-Based HDAC Inhibitors through Alteration of Prodrug Properties. ACS Med Chem Lett5:905-10 (2014) [PubMed] Article