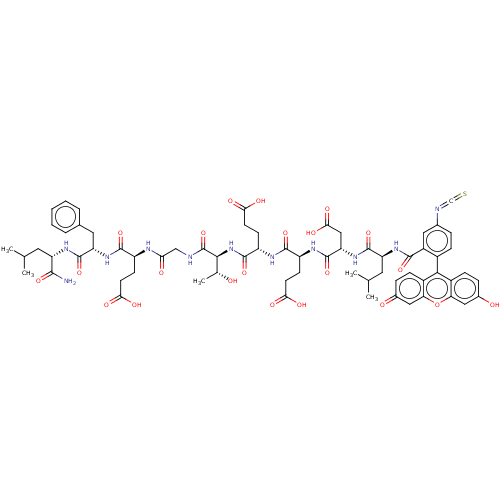
| SMILES | CC(C)C[C@H](NC(=O)[C@H](Cc1ccccc1)NC(=O)[C@H](CCC(O)=O)NC(=O)CNC(=O)[C@@H](NC(=O)[C@H](CCC(O)=O)NC(=O)[C@H](CCC(O)=O)NC(=O)[C@H](CC(O)=O)NC(=O)[C@H](CC(C)C)NC(=O)c1cc(ccc1-c1c2ccc(O)cc2oc2cc(=O)ccc12)N=C=S)[C@@H](C)O)C(N)=O |r,wU:62.63,45.46,32.33,95.101,19.20,8.8,4.3,wD:54.55,36.37,(-37.03,10.94,;-35.96,10.31,;-34.89,10.92,;-35.97,8.77,;-34.65,7.99,;-34.66,6.45,;-33.33,5.67,;-32.26,6.28,;-33.34,4.13,;-34.68,3.38,;-36.01,4.16,;-35.99,5.7,;-37.32,6.48,;-38.66,5.72,;-38.67,4.18,;-37.35,3.4,;-32.01,3.35,;-30.67,4.11,;-30.66,5.34,;-29.34,3.33,;-29.36,1.79,;-30.7,1.03,;-30.72,-.51,;-29.66,-1.13,;-31.79,-1.11,;-28,4.09,;-26.68,3.31,;-26.69,2.08,;-25.33,4.07,;-24.01,3.28,;-22.67,4.04,;-22.66,5.27,;-21.34,3.26,;-20,4.02,;-18.67,3.24,;-18.68,2.01,;-17.33,4,;-17.32,5.54,;-18.65,6.32,;-18.64,7.86,;-17.57,8.46,;-19.7,8.48,;-16,3.22,;-14.66,3.97,;-14.65,5.2,;-13.34,3.19,;-13.35,1.65,;-14.69,.9,;-14.71,-.64,;-13.65,-1.27,;-15.78,-1.25,;-11.99,3.95,;-10.67,3.17,;-10.68,1.94,;-9.33,3.93,;-9.32,5.47,;-10.64,6.25,;-11.72,5.64,;-10.64,7.48,;-8,3.15,;-6.66,3.9,;-6.65,5.14,;-5.33,3.12,;-5.35,1.58,;-6.69,.83,;-6.7,-.4,;-7.75,1.45,;-3.99,3.88,;-2.66,3.1,;-2.67,2.07,;-1.32,3.86,;-1.31,5.4,;.03,6.16,;1.35,5.38,;1.34,3.84,;0,3.08,;,1.54,;-1.33,.77,;-2.67,1.54,;-4,.77,;-4,-.77,;-5.07,-1.39,;-2.67,-1.54,;-1.33,-.77,;,-1.54,;1.31,-.77,;2.67,-1.54,;4,-.77,;5.07,-1.39,;4,.77,;2.67,1.54,;1.31,.77,;.03,7.7,;-1.3,8.48,;-2.36,9.1,;-21.36,1.72,;-20.3,1.09,;-22.43,1.12,;-33.31,8.76,;-32.25,8.14,;-33.3,9.99,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Jain, AD; Potteti, H; Richardson, BG; Kingsley, L; Luciano, JP; Ryuzoji, AF; Lee, H; Krunic, A; Mesecar, AD; Reddy, SP; Moore, TW Probing the structural requirements of non-electrophilic naphthalene-based Nrf2 activators. Eur J Med Chem103:252-68 (2015) [PubMed] Article
Jain, AD; Potteti, H; Richardson, BG; Kingsley, L; Luciano, JP; Ryuzoji, AF; Lee, H; Krunic, A; Mesecar, AD; Reddy, SP; Moore, TW Probing the structural requirements of non-electrophilic naphthalene-based Nrf2 activators. Eur J Med Chem103:252-68 (2015) [PubMed] Article